Abstract
The activity of transcription factors modulates several neural pathways that mediate complex behaviors. We describe here the role of the POU transcription factor UNC-86 in the olfactory behavior of Caenorhabditis elegans. unc-86-null mutants are defective in response to odor attractants but avoid odor repellents normally. Continuous UNC-86 activity is necessary for maintenance of odortaxis behavior; hyperactivation of UNC-86 by fusion to a VP16 activation domain dramatically enhances sensitivity to odor attractants and promotes odor-attractant adaptation. UNC-86 is not expressed in olfactory sensory neurons but is expressed throughout the life of the animal in the AIZ interneurons of the odorsensory pathway. We suggest that UNC-86 transcriptional activity regulates the expression of genes that mediate synaptic properties of AIZ and that hyperactive UNC-86::VP16 may enhance the expression of synaptic components to affect the capacity to analyze and process sensory information.
Odor sensation involves a neural pathway that translates the perception of volatile compounds into motor outputs such as attraction, avoidance, feeding, and mating. To generate such behaviors, an animal recognizes odors, integrates temporal and spatial changes in sensory information, and adjusts its movement. The olfactory system allows animals to sense potential food, danger, and mates. Influenced by previous odorant exposure and other experiences, the sensitivity of an animal to a given odorant may vary over a wide range. Although signal-transduction cascades in olfactory sensory neurons have been explored extensively (1, 2) and molecular mechanisms for plasticity of sensory signaling pathways are beginning to emerge (3–5), neural pathways that integrate and relay odorant stimuli to motor outputs and the mechanisms of behavioral modulation are largely unknown.
Odor sensation and the neurons that mediate olfactory behaviors have been analyzed in Caenorhabditis elegans. Odorants stimulate one of two alternative responses: taxis (attraction) or avoidance. Five pairs of olfactory sensory neurons sense volatile chemicals; each pair detects distinct odorants (6). For example, the AWA neurons detect the attractants diacetyl and pyrazine, the AWC neurons detect the attractants benzaldehyde and isoamyl alcohol, and the AWB neurons detect the repellent nonanone (7). The olfactory sensory neurons extend their dendrites to the tip of the nose and project their axons to the nerve ring (the C. elegans CNS equivalent), where they synapse extensively to the interneurons AIY and AIZ, which connect to command interneurons and motor neurons (8). Neural connectivity, genetic analysis (9, 10), and laser-ablation experiments (7) all implicate the AIZ interneurons as a prominent integrating center that couples odorsensory signals to the behavioral response. After continuous exposure to an odorant, C. elegans displays diminished response to the odorant. This olfactory-adaptation behavior is modulated by the availability of food (11, 12).
The POU transcription factor UNC-86 is expressed specifically in 22 neural lineages during development and in 57 neurons throughout the life of the animal, including the AIZ interneurons (10). unc-86-null mutant animals are fully viable but display odorsensory, chemosensory, and mechanosensory defects (10, 13). The role of UNC-86 in the mechanosensory neurons has been best characterized: UNC-86 specifies neuroblast cell fates to generate the mechanosensory neurons where UNC-86 continuously regulates the expression of the LIM-homeobox gene mec-3 (14–16). UNC-86 also interacts with MEC-3: an UNC-86–MEC-3 protein complex regulates the expression of touch-receptor neuron-specific components, such as mec-7 β-tubulin and mec-4 ion channel (17) to maintain the mechanosensory function of these neurons. Mammalian unc-86 orthologs Brn-3a and Brn-3b also mediate the generation and maintenance of mechanosensory neurons (18) and regulate the expression of axon-guidance genes (19, 20). These studies indicate that transcription factors acting early in the pathway of neural specification may continue to function in the differentiated neurons. The detailed dissection of UNC-86 action in the mechanosensory neurons shows that UNC-86 may interact with cell-specific cofactors to regulate neuron-specific components in each neuron type.
Homeobox genes have been shown to regulate neuronal signaling. The mouse paired-homeobox gene Crx-1 regulates the expression of photoreceptor-signaling components (21, 22), and the hox-like homeodomain protein IDX1/IPF1 regulates the expression of the somatostatin neuropeptide gene (23). C. elegans unc-4 regulates the expression of synaptic components and specifies synaptic connectivity (24, 25). Drosophila Cfla and islet (26, 27) and C. elegans unc-30 (28, 29) regulate neurotransmitter production and packaging in particular neurons. There is evidence from many systems that the expression of synaptic components is modulated by neural activity and that changes in synaptic strength underlie neural and behavioral plasticity (30–32). An outstanding question is whether the activity of transcription factors that trigger cascades of synaptic-component gene expression during initial neurogenesis is also regulated by neural activity in mature neurons to mediate changes in synaptic strength.
Here we report the effect of modulating UNC-86 transcriptional activity on the olfactory behavior of C. elegans. We show that unc-86 loss-of-function mutations impair odor-attractive behavior but leave odor-repellent responses intact. We found that continuous unc-86 activity is necessary for the maintenance of odortaxis behavior. Furthermore, wild-type animals bearing a fusion of UNC-86 to a VP16 transcriptional-activation domain (33) show increased sensitivity to odor attractants and accelerated odor-attractant adaptation. unc-86 is not expressed in the odorsensory neurons that detect odors but is expressed in AIZ, the major interneuron of the olfactory pathway. We suggest that UNC-86 transcriptional activity in AIZ is normally regulated to modulate olfactory acuity and that unc-86::VP16 constitutively activates unc-86-regulated cascades and enhances the synaptic efficacy of the olfactory neural circuit.
Materials and Methods
Constructs. The construction of unc-86(+) and unc-86::VP16 and the generation of transgenic C. elegans that carry the constructs have been described (34). The individual constructs were injected into lin-15(n765) animals; the plasmids containing a mec-7::GFP and the wild-type lin-15 gene, respectively, were coinjected as transformation markers. The individual transgene array was integrated into a chromosome and backcrossed four times with wild-type animals.
To test unc-86::VP16 in other mutant backgrounds, an integrated unc-86::VP16 transgene was crossed into various mutants. In each case, the animals carrying homozygous mutations and a heterozygous mec-7::GFP marker were first identified. Their progeny without the mec-7::GFP marker should show 100% mutant phenotypes. The progeny carrying homozygous mec-7::GFP were then isolated and tested.
Odortaxis Assays. All of the odor assays were performed with well fed young-adult animals as described (7). Because old unc-86 and lin-11 mutant adults carry many fertilized eggs in the uterus and do not move well, only young adults were assayed.
Unless specified, the dilutions of odorants in ethanol were 1:200 benzaldehyde, 1:200 isoamyl alcohol, 1:1,000 diacetyl, undiluted 2-nonanone. The odortaxis index was calculated as [(number of animals at odorant) - (number of animals at solvent ethanol)]/(total number of animals). The odor-avoidance index was calculated as [(number of animals moving from the starting position to the direction opposite the odorant) - (number of animals moving toward the odorant)]/(total number of animals moved).
The odorant-adaptation assays were performed following the procedure described by Colbert and Bargmann (11). The error bars in the figures represent the SEM. The statistical significance of the odortaxis behavior of unc-86(n848ts) mutants from various growth temperatures was determined by using one-way ANOVA.
GFP Expression and Immunoanalysis. The lin-11::GFP was provided by O. Hobert (Columbia University, New York). The anti-UNC-86 antibody immunostaining followed the procedures of Finney and Ruvkun (10).
Results
unc-86-Null Mutations Affect Odor-Attraction but Not Odor-Avoidance Behavior. Of the neurons involved in the olfactory behavior of C. elegans, the development of the AIZ interneurons requires the POU-homeodomain transcription factor UNC-86. The pair of bilaterally symmetric AIZ interneurons is generated from a stereotyped cell lineage during late embryogenesis (35). Each AIZ neuron extends a single axon to the nerve ring, where it receives synaptic inputs from many odorsensory and chemosensory neurons; the AIZ neurons have synaptic outputs to command interneurons and motor neurons (ref. 8 and Fig. 1a). The expression of UNC-86 is initiated in the neuroblasts that generate AIZ; unc-86-null mutations cause a lineage transformation of the neuroblasts so that the AIZ interneurons are not generated (refs. 9 and 10 and Fig. 1b). Consistent with these data, unc-86 mutants have olfactory defects (7, 10), but detailed behavioral deficits have not been reported.
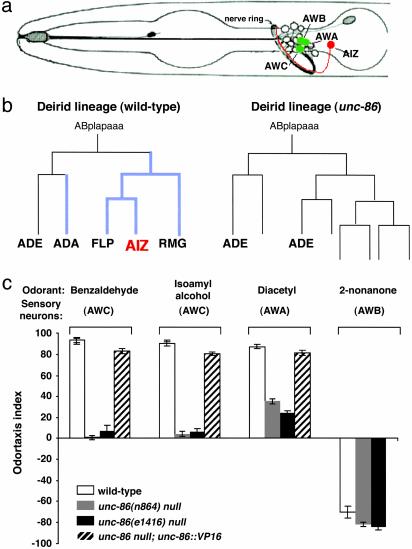
Fig. 1.
Effects of unc-86-null mutations on the olfactory system. (a) A schematic representation of the position of the AIZ interneurons and the olfactory sensory neurons AWA, AWC, and AWB. Each type of neuron is a bilateral symmetric pair; a lateral view is shown. The drawing is adapted from Starich et al. (36). (b) AIZ are the only neurons in the odor-sensation neuronal pathway affected by unc-86 mutations. UNC-86 expression is initiated in the neuroblasts of the lineage (10). The unc-86-null mutation transforms the neuroblast cell fates and AIZ are not generated (9). Cells expressing UNC-86 are indicated by thickened lines. (c) The unc-86-null mutants n846 and e1416 are defective in response to odor attractants, but respond as well as wild-type animals to odor repellents. The unc-86::VP16 fusion transgene can restore unc-86-null mutant odor-attractant responses.
We assayed the olfactory behavior of two unc-86-null mutant strains, unc-86(n846) and unc-86(e1416). Well-fed adult wild-type and unc-86-null animals were compared for their responses to a set of previously established odorants (7). Whereas ≈90% of wild-type animals migrate to the source of odor attractants, unc-86-null mutant animals fail to respond to the AWC-detected attractants benzaldehyde and isoamyl alcohol and have ≈30% normal sensitivity to the AWA-detected attractant diacetyl (Fig. 1c). In contrast, the unc-86-null mutant animals respond as well as wild-type animals to the AWB-detected repellent 2-nonanone (Fig. 1c). unc-86-null mutant animals also display normal responses to the ADL- and ASH-detected repellent 1-octanol (refs. 6 and 37 and data not shown). Thus, unc-86-null mutant animals have the ability to move in such a behavioral assay, but are defective specifically in the response to odor attractants.
UNC-86 is not expressed in odorsensory or chemosensory neurons, but is expressed in the AIZ interneurons (10). AIZ are a major postsynaptic target of the AWA and AWC odor-attractant sensory neurons and are important for olfactory behavior (7). Thus, it is likely that the olfactory behavior defects in the unc-86-null mutant are due to missing AIZ-mediated odor-sensory signal integration and processing. Because the unc-86-null mutant animals respond normally to odor repellents, AIZ neurons are not required for odor-avoidance behavior.
unc-86 Activity Is Necessary for Maintenance of AIZ Interneuron Function. The expression of unc-86 in AIZ is sustained throughout the life of the animal (ref. 10, Fig. 2a), which raises the possibility that unc-86 may also function in the maintenance of AIZ signaling. We explored this role of UNC-86 by analyzing odortaxis behavior of the temperature sensitive unc-86(n848ts) mutant (13).
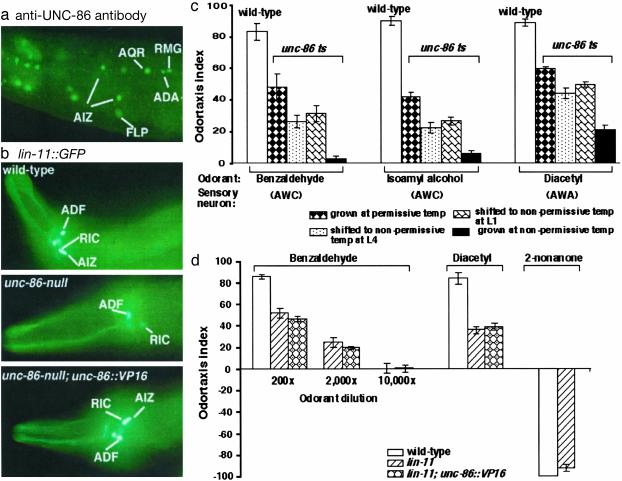
Fig. 2.
UNC-86 is required for AIZ function and the maintenance of odortaxis behavior. (a) UNC-86 is expressed continuously in the pair of AIZ interneurons throughout the life of the animal as detected by immunostaining with anti-UNC-86 antibodies. Anterior is on the left. (b) UNC-86 regulates the expression of the LIM-factor lin-11 in AIZ. In wild-type animals, the lin-11::GFP is expressed in the AIZ interneurons as well as several other head neurons. unc-86-null animals fail to express lin-11::GFP in AIZ, whereas expression in other neurons is normal. unc-86-null animals carrying an integrated unc-86::VP16 fusion gene show a wild-type lin-11::GFP expression in AIZ and the other neurons. All strains carry the same integrated lin-11::GFP fusion gene. The animals are young adults. Anterior is on the left. (c) UNC-86 is necessary for the maintenance of olfactory behavior. Wild-type and the unc-86(n848ts) temperature-sensitive mutants grown under the same conditions were compared for response to odor attractants. The animals were kept at 15°C, shifted to the nonpermissive temperature (25°C) at indicated developmental stages, and assayed at the adult stage. unc-86(n848) animals grown at the nonpermissive temperature exhibit odortaxis deficits equivalent to unc-86-null mutants (Fig. 1). unc-86(n848) animals shifted to the nonpermissive temperature at larval stages show more profound odortaxis defects than those grown at the permissive temperature (P < 0.003 for benzaldehyde and P < 0.007 for isoamyl alcohol), but there is no significant difference between unc-86(n848) animals shifted to the nonpermissive temperature at larval stage 1 (L1) or larval stage 4 (L4). Changes of temperature do not affect odortaxis behaviors of wild-type animals; odortaxis behaviors of wild type grown at 25°C are shown. (d) Deletion of the LIM-factor gene lin-11 affects odortaxis behavior. Two lin-11 alleles (n566 and n1281) were assayed. Both alleles show similar behavioral defects, lin-11(n566) results are shown. Mutations of lin-11 suppress unc-86::VP16-directed odor-attractant hypersensitivity, indicating the action of unc-86::VP16 requires the lin-11 gene function.
The AIZ interneurons are born during embryogenesis. They can be identified in living animals based on the characteristic position of the cell body and by using the expression of GFP fusion to the LIM-homeobox gene lin-11 (Fig. 2b). The AIZ neurons are the only cells that express both lin-11 and unc-86 (10, 38). unc-86(n848ts) animals grown at the nonpermissive temperature (25°C) display olfactory defects equivalent to unc-86-null mutants (Fig. 2c) and do not express lin-11::GFP in AIZ (n > 50), whereas wild-type animals grown at the same temperature respond to odor attractants normally (Fig. 2c) and express lin-11::GFP in AIZ throughout the life of the animal. unc-86(n848) animals grown at the permissive temperature (15°C) show lin-11::GFP expression in AIZ (n = 155) but display partial odor attraction (Fig. 2c), suggesting that unc-86(n848) at the permissive temperature provides sufficient unc-86 activity for at least some AIZ properties and partial perception of odor attractants.
The unc-86(n848ts) mutants shifted to the nonpermissive temperature at larval stages (after the AIZ generation) display more profound odor-attractant behavioral defects when tested as adults (Fig. 2c). This finding indicates that UNC-86 acts after the generation of AIZ to regulate odortaxis behavior. Similar deficits in odortaxis were observed in unc-86(n848ts) adults shifted to the nonpermissive temperature at larval stage 1 or larval stage 4 (Fig. 2c), suggesting that continuous UNC-86 activity is needed to sustain odortaxis behavior.
lin-11 regulates AIZ axonal morphology (38). The lin-11(n566) mutation is a probable null allele (39). lin-11(n566) animals are defective in response to both AWA- and AWC-mediated odor attractants, but respond normally to 2-nonanone (Fig. 2d). The coexpression of UNC-86 and LIN-11 in AIZ and the similar pattern of olfactory defects suggest that lin-11 and unc-86 are both likely to act in AIZ to regulate olfactory behavior. lin-11-null mutants are not as defective in odortaxis as unc-86-null mutants, which suggests either that AIZ are partially functional in the lin-11-null mutant or that other interneurons not affected by lin-11 but affected by unc-86 also transduce signaling from AWA and AWC. It is probably a significant distinction that AIZ are generated in the lin-11-null mutant but not in the unc-86-null mutant (9, 38).
Activation of unc-86 Transcriptional Activity Enhances Olfactory Sensitivity. We addressed whether the activity of the UNC-86 transcription factor might modulate the olfactory behavior of C. elegans. To activate UNC-86, we fused the strong transcriptional-activation domain from the herpes virus VP16 protein to UNC-86 (34). We reasoned that if UNC-86 transcriptional activity in AIZ is normally coupled to neural activity, fusion to the potent VP16 activation domain might disturb finely regulated expression of unc-86 downstream genes and, thus, modify AIZ synaptic properties and cause behavioral changes.
The VP16 activation domain was inserted into a nonconserved coding region upstream of the POU domain in an unc-86 genomic fragment. This fusion gene is expressed congruently with chromosomal unc-86 (34). We generated transgenic animals carrying the unc-86::VP16 fusion gene integrated into a chromosome and observed the effect on the AIZ development and odortaxis behavior. unc-86-null mutants carrying the unc-86::VP16 fusion gene generate the AIZ interneurons normally (Fig. 2b) and restore odortaxis behavior (Fig. 1c). Thus, the unc-86::VP16 fusion gene can replace unc-86(+) for the specification of the neuroblast to generate AIZ, as well as for the odortaxis behavior.
Interestingly, wild-type animals expressing the unc-86::VP16 fusion protein show markedly increased odor-attractant sensitivity (Fig. 3 a and b). At high concentrations of benzaldehyde, both wild-type and unc-86::VP16 animals respond vigorously. When benzaldehyde is serially diluted, however, unc-86::VP16 animals are much more responsive than wild-type animals (Fig. 3a). This enhanced odor sensitivity is dependent on the VP16 transactivation domain, because wild-type animals that carry an integrated unc-86 transgene [unc-86(+)] exhibit a similar odor sensitivity to wild-type animals (Fig. 3a). unc-86::VP16 animals are also more sensitive than wild type to the attractants isoamyl alcohol and diacetyl, although the effect of unc-86::VP16 on the diacetyl sensitivity is less pronounced (Fig. 3 b and c). Consistent with the normal odor-repellent behavior of unc-86-null mutant animals, unc-86::VP16 has no effect on the sensitivity to the AWB-detected repellent nonanone (Fig. 3d). This normal behavior on an odor-avoidance test also shows that the increased activity in the odor-attraction experiments is not simply due to hyperactivity of the animals.
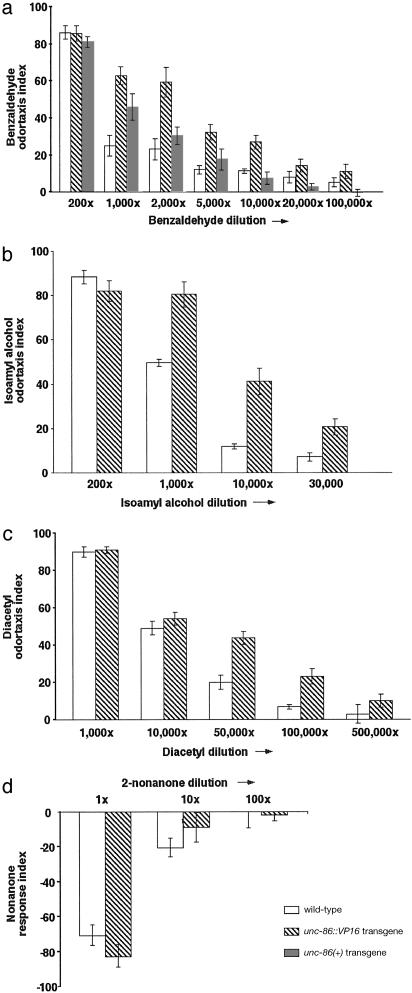
Fig. 3.
Transgenic wild-type animals that carry an integrated unc-86::VP16 fusion gene have increased sensitivity to odor attractants. Wild-type and transgenic animals that carry unc-86(+)or unc-86::VP16 transgene were compared for the response to serial diluted benzaldehyde detected by AWC (a), isoamyl alcohol detected by AWC (b), diacetyl detected by AWA (c), and 2-nonanone detected by AWB (d). unc-86(+) and the unc-86::VP16 transgene carry the identical unc-86 sequences; the unc-86::VP16 fusion gene has the activation domain of the viral protein VP16 inserted at a nonconserved region of the unc-86(+) sequence (34). Two independent unc-86::VP16 integrant lines were assayed; the summary of one line is shown.
The increased sensitivity to odorants conferred by unc-86::VP16 is dependent on the normal olfactory sensory neurons. To determine whether unc-86::VP16 activity alters sensory specificity, we crossed the unc-86::VP16 fusion gene into mutants defective specifically in either the AWA or AWC olfactory neuron function and tested if unc-86::VP16 can improve the response to defects in AWA- or AWC-mediated odor attractants, respectively. odr-7 encodes a transcription factor that is necessary for AWA function and for the expression of the odr-10 diacetyl receptor (40, 41). The odr-7(ky4) animals bearing the unc-86::VP16 transgene show no improvement in diacetyl sensation (Fig. 4). odr-1 encodes a guanylyl cyclase that is required for AWC- but not AWA-mediated olfaction (42). odr-1 mutants carrying unc-86::VP16 exhibit the same benzaldehyde sensory defects as odr-1 mutants (Fig. 4). Thus, the normal sensory input from the AWC and AWA sensory neurons is required in unc-86::VP16 animals. A lin-11(n566) mutation suppresses the unc-86::VP16-induced hypersensitivity to odor attractants (Fig. 2d), showing that the unc-86::VP16-induced olfactory hypersensitivity depends on the lin-11 gene activity.
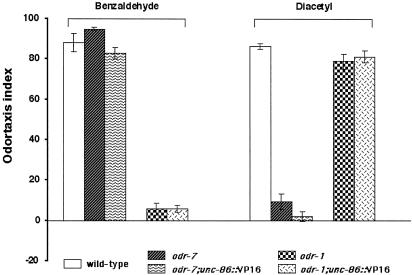
Fig. 4.
unc-86::VP16 does not alter sensory neuron AWA and AWC odorant specificity. The ODR-7 nuclear receptor is necessary for the expression of the putative diacetyl receptor ODR-10 in AWA; odr-7(ky4) is a null mutant (40, 41). Both odr-7 and odr-7; unc-86::VP16 strains failed to respond to diacetyl. odr-1 acts in AWC but not AWA (42); both odr-1 and odr-1; unc-86::VP16 animals do not respond AWC-detected odorants.
Together, these results suggest that unc-86::VP16 plays the same role as unc-86 in the olfactory neural circuit and that activation of UNC-86 by the VP16 transcription domain enhances the perception of the odor-attractant intensity.
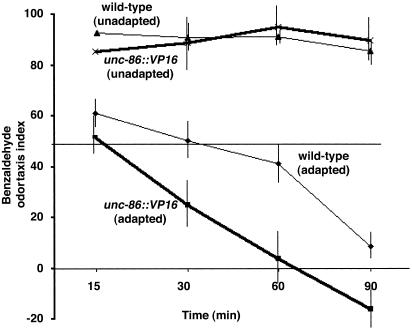
unc-86::VP16 Induces Accelerated Olfactory Adaptation. Odor sensation in C. elegans involves not only the discrimination of volatile molecules, but also the modification of the behavioral repertoire based on previous exposure to odorant stimuli (11, 12). Continuous exposure to an odor attractant causes a decline in the response to that odor attractant. This desensitization or adaptation odor-response behavior depends on the time and intensity of the odor stimuli (11). We tested if unc-86::VP16 also modifies odor-attractant adaptation behavior. After continuous exposure to a high concentration of the odor attractant benzaldehyde, both wild-type and unc-86::VP16 transgenic animals displayed a decreased response to benzaldehyde. unc-86::VP16 animals, however, adapt to benzaldehyde ≈2–2.5 times faster than wild-type animals (Fig. 5). This observation indicates that unc-86::VP16 animals, like wild-type animals, can adjust their sensitivity based on previous experience, but that unc-86::VP16 enhances the speed of olfactory adaptation.
Fig. 5.
unc-86::VP16 promotes odor-attractant adaptation. Wild-type and unc-86::VP16 animals were exposed continuously to concentrated benzaldehyde for a period as indicated and were then tested for attraction to benzaldehyde.
Discussion
unc-86 Is Necessary for Maintenance of Odortaxis Behavior. These results show that the POU-homeodomain transcription factor UNC-86 acts in mature animals to regulate olfactory behavior. By using the temperature-sensitive unc-86(n848ts) mutation, we show that continuous unc-86 gene activity is necessary to maintain odortaxis behavior of the adult animal (Fig. 2c). unc-86-null mutants are defective in the response to both AWA- and AWC-mediated odorants, but respond normally to odorants detected by AWB, ADL, and ASH. This finding indicates that unc-86 regulates the neural circuits specifically coupling sensory signals and odortaxis behavior. There is evidence that it is the sensory neurons that dictate olfactory response in C. elegans; for example, animals avoid diacetyl if the diacetyl receptor odr-10, normally expressed in AWA, is expressed in AWB but not AWA (43). Together, these studies suggest that odor-attraction and -avoidance behavior in C. elegans is specified by distinct synaptic connectivity between the olfactory sensory neurons and their targets. In mammals, the topographic organization of the olfactory sensory neurons and their projections to the glomeruli in the olfactory bulb specify the quality of odorants; each glomerulus serves as a functional unit to translate the signals detected by the connected olfactory sensory neurons to distinct olfactory perception (44). Thus, some principles of olfactory discrimination in the neural circuits are conserved between worms and mammals.
unc-86 is not expressed in any of the olfactory sensory neurons, but it is expressed in the AIZ interneurons (10). Several lines of evidence suggest that UNC-86 is likely to act in the AIZ interneurons to regulate odortaxis behavior. First, of 57 neurons that express UNC-86, only AIZ have been implicated in olfactory behavior (7); AIZ are not generated in unc-86 mutants. Second, unc-86 activity regulates the expression of LIM-factor lin-11 in AIZ, and lin-11 mutants show odortaxis defects but normal odor avoidance behavior. lin-11 is also expressed in the AWA neurons (45), which could contribute to the defects in the response to diacetyl that is detected by AWA. Because lin-11 is not expressed in AWC, the defects in response to benzaldehyde and isoamyl alcohol could be caused by defective AIZ in lin-11 mutants. lin-11 may function in the AIZ interneurons analogously to the LIM gene mec-3 in the mechanosensory neurons. unc-86 activates mec-3 expression for the touch-receptor neuron differentiation and function (14). The interaction between UNC-86 and MEC-3 increases DNA binding affinity and specificity and is necessary for mec-7 β-tubulin and mec-4 ion-channel gene expression (17). Thus, UNC-86 may regulate different target genes by interacting with neuron-specific cofactors to control specific behavior. lin-11 is coexpressed with unc-86 only in the AIZ neurons, so we expected that expression of unc-86 coding region from a lin-11 promoter might rescue the chemosensory defects of unc-86. We were not able, however, to rescue unc-86(n848ts) odortaxis defects by expressing unc-86 coding region under the control of a lin-11 promoter (unpublished observations). It is possible that some UNC-86-regulated genes normally expressed earlier in the lineage cannot be rescued by the late expression of unc-86 from the lin-11 promoter. It is also possible that UNC-86-expressing cells in addition to AIZ have a role in the olfactory neural circuit.
Activation of UNC-86 Causes Hypersensitivity to Odor-Attractant Stimulation. Transcriptional regulation of signaling and synaptic components has been found to couple neural activity with long-term changes in synaptic strength (32, 46, 47). The transcription factor CREB, for example, is modulated by phosphorylation to couple a variety of physiological stimuli to the differential expression of downstream genes that mediate synaptic and behavioral plasticity (48–50).
UNC-86 fused to the VP16 transcriptional-activation domain can replace wild-type unc-86 to direct odortaxis behavior, but animals bearing the unc-86::VP16 fusion gene respond better than wild-type animals to low levels of odor attractants (Fig. 3). This result is opposite to the reduced odor attractant sensitivity caused by reduction of unc-86 activity and indicates that unc-86::VP16 activity enhances olfactory acuity.
C. elegans responds to many environmental stimuli, including odor, temperature, food, and pheromone; many of these inputs interact to affect behaviors (6). The AIZ interneurons are a likely site of such sensory-behavioral integration. AIZ is connected to multiple sensory inputs, and animals missing AIZ because of either laser ablation or mutations show abnormal responses to many sensory stimuli (6). POU-domain factors regulate synapse formation and synaptic transmission. For example, the unc-86 mammalian orthologue Brn-3 subfamilies regulate the expression of the acetylcholine receptor and the synaptic vesicular protein SNAP-25 (19, 20, 51). It is possible that UNC-86 in the mature AIZ neurons mediates the maintenance of signaling components, such as neurotransmitters, receptors, and synaptic components; it is also possible that the transcriptional activity of UNC-86 is coupled to sensory neural activity. UNC-86 bears potential phosphorylation sites for kinases that are regulated by neural activity, such as PKA, PKC, and CK2. Like CREB and FOS, the transcriptional-activation function of UNC-86 may be modulated by phosphorylation. Constitutive activation of UNC-86 by fusion of the VP16 transcriptional-activation domain may increase or misregulate the expression of these signaling molecules and impose lasting changes in the synaptic strength among AIZ and their pre- or postsynaptic partners, reminiscent of transcription-dependent LTP at synapses induced by repeated stimuli and of activated CREB-induced modification of synaptic strength (48).
unc-86 also regulates serotonin synthesis in a subset of C. elegans serotonergic neurons (52). Serotonin signaling mediates many food-modulated behaviors (53). Brief starvation enhances olfactory adaptation, and application of exogenous serotonin can suppress this starvation-induced olfactory-adaptation facilitation (12). The unc-86::VP16 enhancement of olfactory adaptation could be interpreted as an indirect effect of induction of starvation signals. This model would predict that unc-86::VP16 decreases serotonin production to cause olfactory behavioral changes. In wild-type animals, however, unc-86 activates serotonin synthesis in serotonergic neurons and unc-86::VP16 does not affect serotonin synthesis in head neurons (ref. 52; unpublished observations), suggesting that alterations in serotonin signaling are unlikely the cause of increased olfactory adaptation. Rather, we favor the hypothesis that UNC-86::VP16-induced expression of unc-86 downstream genes in AIZ, which are connected to serotonergic neurons (8), promotes odor-attractant adaptation even in the absence of starvation signals. Further molecular analysis is necessary to understand which neuronal components in AIZ are regulated by unc-86 and how UNC-86 protein activity is modulated by sensory neural activity.
Acknowledgments
We thank Dr. Catherine Dempsey for statistical analysis of behavioral results and Yanxia Liu for microinjection. J.Y.S. was supported by a National Institute of Neurological Disorders and Stroke fellowship. G.R. was supported by a Hoechst AG grant.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Prasad, B. C. & Reed, R. R. (1999) Trends Genet. 15, 150-153. [DOI] [PubMed] [Google Scholar]
- 2.Ronnett, G. V. & Moon, C. (2002) Annu. Rev. Physiol. 64, 189-222. [DOI] [PubMed] [Google Scholar]
- 3.Kurahashi, T. & Menini, A. (1997) Science 385, 725-729. [DOI] [PubMed] [Google Scholar]
- 4.Zufall, F. & Leinders-Zufall, T. (2000) Chem. Senses 25, 473-481. [DOI] [PubMed] [Google Scholar]
- 5.L'Etoile, N. D., Coburn, C. M., Eastham, J., Kistler, A., Gallegos, G. & Bargmann, C. I. (2002) Neuron 36, 1079-1089. [DOI] [PubMed] [Google Scholar]
- 6.Bargmann, C. I. & Mori, I. (1997) in C. elegans II, eds. Riddle, D. L., Blumenthal, T., Meyer, B. J. & Priess, J. R. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 717-737.
- 7.Bargmann, C. I., Hartwieg, E. & Horvitz, H. R. (1993) Cell 74, 515-527. [DOI] [PubMed] [Google Scholar]
- 8.White, J. G., Southgate, E., Thomson, J. N. & Brenner, S. (1986) Philos. Trans. R. Soc. London B 314, 1-340. [DOI] [PubMed] [Google Scholar]
- 9.Chalfie, M., Horvitz, H. R. & Sulston, J. E. (1981) Cell 24, 59-69. [DOI] [PubMed] [Google Scholar]
- 10.Finney, M. & Ruvkun, G. (1990) Cell 63, 895-905. [DOI] [PubMed] [Google Scholar]
- 11.Colbert, H. A. & Bargmann, C. I. (1995) Neuron 14, 803-812. [DOI] [PubMed] [Google Scholar]
- 12.Colbert, H. A. & Bargmann, C. I. (1997) Learn. Mem. 4, 179-191. [DOI] [PubMed] [Google Scholar]
- 13.Finney, M., Ruvkun, G. & Horvitz, H. R. (1988) Cell 55, 757-769. [DOI] [PubMed] [Google Scholar]
- 14.Way, J. C. & Chalfie, M. (1989) Genes Dev. 3, 1823-1833. [DOI] [PubMed] [Google Scholar]
- 15.Xue, D., Finney, M., Ruvkun, G. & Chalfie, M. (1992) EMBO J. 11, 4969-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue, D., Tu, Y. & Chalfie, M. (1993) Science 261, 1324-1328. [DOI] [PubMed] [Google Scholar]
- 17.Duggan, A., Ma, C. & Chalfie, M. (1998) Development (Cambridge, U.K.) 125, 4107-4119. [DOI] [PubMed] [Google Scholar]
- 18.McEvilly, R. J. & Rosenfeld, M. G. (1999) Prog. Nucleic Acid Res. Mol. Biol. 63, 223-255. [DOI] [PubMed] [Google Scholar]
- 19.Smith, M. D., Dawson, S. J. & Latchman, D. S. (1997) Mol. Cell. Biol. 17, 345-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erkman, L., Yates, P. A., McLaughlin, T., McEvilly, R. J., Whisenhunt, T., O'Connell, S. M., Krones, A. I., Kirby, M. A., Rapaport, D. H., Bermingham, J. R., et al. (2000) Neuron 28, 779-792. [DOI] [PubMed] [Google Scholar]
- 21.Furukawa, T., Morrow, E. M. & Cepko, C. L. (1997) Cell 91, 531-541. [DOI] [PubMed] [Google Scholar]
- 22.Chen, S., Wang, Q., Nie, Z., Sun, H., Lennon, G., Copeland, N. G., Gilbert, D. J., Jenkins, N. A. & Zack, D. J. (1997) Neuron 19, 1017-1030. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz, P. T., Perez-Villamil, B., Rivera, A., Moratalla, R. & Vallejo, M. (2000) J. Biol. Chem. 275, 19106-19114. [DOI] [PubMed] [Google Scholar]
- 24.Miller, D. M., Shen, M. M., Shamu, C. E., Burglin, T. R., Ruvkun, G., Dubois, M. L., Ghee, M. & Wilson, L. (1992) Nature 355, 841-845. [DOI] [PubMed] [Google Scholar]
- 25.Lickteig, K. M., Duerr, J. S., Frisby, D. L., Hall, D. H., Rand, J. B. & Miller, D. M, III (2001) J. Neurosci. 21, 2001-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, W. A. & Hirsh, J. (1990) Nature 343, 467-470. [DOI] [PubMed] [Google Scholar]
- 27.Thor, S. & Thomas, J. B. (1997) Neuron 18, 397-409. [DOI] [PubMed] [Google Scholar]
- 28.McInitre, S. L., Jorgensen, E. & Horvitz, R. H. (1993) Nature 364, 334-347. [DOI] [PubMed] [Google Scholar]
- 29.Eastman, C., Horvitz, H. R. & Jin, Y. (1999) J. Neurosci. 19, 6225-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, K. C., Barad, M. & Kandel, E. R. (2000) Curr. Opin. Neurobiol. 10, 587-592. [DOI] [PubMed] [Google Scholar]
- 31.Lonze, B. & Ginty, D. (2002) Neuron 35, 605-623. [DOI] [PubMed] [Google Scholar]
- 32.Kandel, E. R. (2001) Biosci. Rep. 21, 565-611. [DOI] [PubMed] [Google Scholar]
- 33.Triezenberg, S. J., Kingsbury, R. C. & McKnight, S. L. (1988) Genes Dev. 2, 718-729. [DOI] [PubMed] [Google Scholar]
- 34.Sze, J. Y., Liu, Y. & Ruvkun, G. (1997) Development (Cambridge, U.K.) 124, 1159-1168. [DOI] [PubMed] [Google Scholar]
- 35.Sulston, J. E., Schierenerg, E., White, J. G. & Thomson, J. N. (1983) Development (Cambridge, U.K.) 100, 64-119. [DOI] [PubMed] [Google Scholar]
- 36.Starich, T. A., Herman, R. K., Kari, C. K., Yeh, W. H., Schackwitz, W. S., Schuyler, M. W., Collet, J., Thomas, J. H. & Riddle, D. L. (1995) Genetics 139, 171-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hart, A., Sims, S. & Kaplan, J. (1995) Nature 378, 82-85. [DOI] [PubMed] [Google Scholar]
- 38.Hobert, O., Liu, Y., D'Alberti, T. & Ruvkun, G. (1998) J. Neurosci. 18, 2084-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freyd, G., Kim, S. K. & Horvitz, H. R. (1990) Nature 344, 876-879. [DOI] [PubMed] [Google Scholar]
- 40.Sengupta, P., Colbert, H. A. & Bargmann, C. I. (1994) Cell 79, 971-980. [DOI] [PubMed] [Google Scholar]
- 41.Sengupta, P., Chou, J. H. & Bargmann, C. I. (1996) Cell 84, 899-909. [DOI] [PubMed] [Google Scholar]
- 42.L'Etoile, N. D. & Bargmann, C. I. (2000) Neuron 25, 575-586. [DOI] [PubMed] [Google Scholar]
- 43.Troemel, E. R., Kimmel, B. E. & Bargmann, C. I. (1997) Cell 91, 161-169. [DOI] [PubMed] [Google Scholar]
- 44.Dulac, C. (1997) Neuron 19, 477-480. [DOI] [PubMed] [Google Scholar]
- 45.Sarafi-Reinach, T. R., Melkman, T., Hobert, O. & Sengupta, P. (2001) Development (Cambridge, U.K.) 128, 3269-3281. [DOI] [PubMed] [Google Scholar]
- 46.West, A. E., Chen, W. G., Dalva, M. B., Dolmetsch, R. E., Kornhauser, J. M., Shaywitz, A. J., Takasu, M. A., Tao, X. & Greenberg, M. E. (2001) Proc. Natl. Acad. Sci. USA 98, 11024-11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dubnau, J. & Tully, T. (2001) Curr. Biol. 11, R240-R243. [DOI] [PubMed] [Google Scholar]
- 48.Yin, J. C. P., Vecchio, M. D., Zhou, H. & Tully, T. (1995) Cell 81, 107-115. [DOI] [PubMed] [Google Scholar]
- 49.Bito, H., Deisseroth, K. & Tsien, R. W. (1996) Cell 87, 1203-1214. [DOI] [PubMed] [Google Scholar]
- 50.Kornhauser, J. M., Cowan, C. W., Shaywitz, A. J., Dolmetsch, R. E., Griffith, E. C., Hu, L. S., Haddad, C., Xia, Z. & Greenberg, M. E. (2002) Neuron 34, 221-233. [DOI] [PubMed] [Google Scholar]
- 51.Milton, N. G. N., Bessis, A., Changeux, J.-P. & Latchman, D. S. (1995) J. Biol. Chem. 270, 15143-15147. [DOI] [PubMed] [Google Scholar]
- 52.Sze, J. Y., Zhang, S., Li, J. & Ruvkun, G. (2002) Development (Cambridge, U.K.) 129, 3901-3911. [DOI] [PubMed] [Google Scholar]
- 53.Weinshenker, D., Garriga, G. & Thomas, J. H. (1995) J. Neurosci. 15, 6975-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]