Abstract
Neural progenitor cells are widespread throughout the adult central nervous system but only give rise to neurons in specific loci. Negative regulators of neurogenesis have therefore been postulated, but none have yet been identified as subserving a significant role in the adult brain. Here we report that nitric oxide (NO) acts as an important negative regulator of cell proliferation in the adult mammalian brain. We used two independent approaches to examine the function of NO in adult neurogenesis. In a pharmacological approach, we suppressed NO production in the rat brain by intraventricular infusion of an NO synthase inhibitor. In a genetic approach, we generated a null mutant neuronal NO synthase knockout mouse line by targeting the exon encoding active center of the enzyme. In both models, the number of new cells generated in neurogenic areas of the adult brain, the olfactory subependyma and the dentate gyrus, was strongly augmented, which indicates that division of neural stem cells in the adult brain is controlled by NO and suggests a strategy for enhancing neurogenesis in the adult central nervous system.
The vast majority of neurons in the mammalian brain are produced during embryonic development. However, remnants of the germinal zones of the developing brain continue to proliferate into adulthood, generating large numbers of neurons in the adult brain (1–3). The subventricular zone (SVZ) of the lateral ventricles (LVs), its anterior extension, the rostral migratory stream (RMS), and the subgranular cell layer (S-GCL) of the dentate gyrus (DG) of the hippocampus are the major sites of adult neurogenesis, although other regions of the adult brain retain the potential to generate new neurons (4–6). Many of the newly generated neurons undergo physiological cell death (7), but it is becoming clear that some of these new neurons become integrated into existing neuronal circuits, thus potentially contributing to a previously unanticipated form of neuroplasticity (8). Several protein growth factors have been shown to affect adult neurogenesis in vivo (5, 6, 9–11). However, the signaling systems involved in regulating cell division in the adult brain are only beginning to be understood.
Increasingly diverse functions of NO, a transcellular signaling molecule (12), are continuing to be demonstrated, and there is growing evidence that NO may be involved in controlling proliferation of neuronal cells. Neuronal NO synthase (nNOS), the major NOS isoform in the mammalian brain, is transiently expressed in the developing brain in a pattern suggesting its involvement in neural development (13). Furthermore, NO has been shown to effectively and reversibly suppress cell division (14, 15); this property of NO, coupled to its ability to regulate gene expression, is exploited in a number of developmental contexts (16).
Materials and Methods
A full description of the methods used in this work can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
All animal procedures were performed under National Institutes of Health guidelines for the care and use of animals in research and approved by the animal care and use committees of the host institutes.
Surgical Procedures and BrdUrd Administration. Osmotic minipumps were used to infuse 50 mM Nω-nitro-methyl-L[D]-arginine methyl ester (NAME) into the LV of Sprague–Dawley rats. BrdUrd was administered i.p. to rats (120 mg/kg) twice daily for 7 days starting on the day after surgery or twice daily for 2.5 days to mice (50 mg/kg).
Histochemistry. At various time points animals were perfused transcardially, and the brains were fixed and sectioned on a cryostat by using standard procedures. Diaphorase reaction and immunohistochemistry were performed by using standard methods. Immunohistochemistry with rabbit anti-nNOS (Z-RNN3, Zymed), anti-BrdUrd antibody [rat ascites, clone BU1/75(ICR1) Harlan Laboratories, Haslett, MI], mouse anti-βIII-tubulin (clone 5G8, Promega), or mouse anti-glial fibrillary acidic protein (clone G-A-5, Roche Molecular Biochemicals) was performed in PBS containing 0.1% triton and 2% normal goat serum. For BrdUrd immunohistochemistry the sections were denatured with acid to expose the BrdUrd epitope. Apoptotic cells were examined by using the terminal deoxynucleotidyltransferase-mediated dUTP end-labeling assay (ApoAlert DNA fragmentation assay kit, CLONTECH). Confocal microscopy was performed on a Zeiss LSM 510 machine by using “multitracking” settings.
Cell Counting. Unbiased absolute counts of BrdUrd-labeled (BrdUrd+) cells were made after peroxidase immunostaining in rats and mice; in rats these were on the hemisphere ipsilateral to the cannula. Counts were made by a blinded observer, and the code was not broken until all the counting was completed. Scoring of BrdUrd/βIII-tubulin-coimmunostained cells was performed by using confocal microscope z series.
Gene Targeting. A null mutant of the mouse nNOS gene [disrupted nNOS allele (KOex6)] carrying a 628-bp-long deletion of exon 6 with adjacent intronic sequences was generated by homologous recombination.
NOS Activity Determination. An assay for the activity of NOS by using the conversion of [3H]arginine to [3H]citrulline was performed according to standard methods.
Results
Localization of nNOS in the Adult Brain Suggests a Role in the Control of Neurogenesis. To examine whether NOS may be involved in the control of adult neurogenesis, we compared sites of neurogenesis in the adult rat brain with the sites of NOS expression (17). We used immunohistochemical detection of nNOS and NADPH-diaphorase histochemical staining for NOS in conjunction with labeling cells in S phase with BrdUrd. Strong diaphorase staining was seen in the cell bodies and the processes of the medium-sized aspiny interneurons in the forebrain and midbrain, in agreement with numerous reports (17, 18). A dense network of diaphorase-positive fibers were seen throughout most parenchymal areas and were particularly dense in the striatum.
Both the SVZ and the RMS neurogenic regions exhibit very little diaphorase staining in comparison with the surrounding parenchyma. In contrast, the distribution of BrdUrd+ cells was limited to the SVZ and RMS, whereas surrounding parenchyma showed very few labeled cells (Fig. 1 a and b, bracketed areas). We confirmed the notion that NOS+ cells are largely excluded from zones of active neurogenesis by staining brain sections with anti-nNOS antibodies (Fig. 1 c and d). Whereas numerous BrdUrd+ cells were seen in the SVZ and the RMS, cell bodies of strongly nNOS+ cells were predominantly located outside of these neurogenic areas (Fig. 1 c and d, arrows). Some of the processes of the nNOS+ cells intermingled with BrdUrd+ cells in the SVZ (Fig. 1 b and c, arrowheads) and RMS.
Fig. 1.
NOS localization in the brain suggests a role for control of neurogenesis. (a and b) Histochemical detection of NADPH-diaphorase (blue staining) and immunochemical detection of BrdUrd (brown staining). Note that there is significantly less diaphorase staining, representing NOS enzyme, in the bracketed areas, which mark the prolific neurogenic regions of the SVZ and RMS. (c and d) Double labeling of nNOS+ (red) and BrdUrd+ (green) cells in the SVZ (c) and RMS (d). Note that bodies of nNOS-expressing neurons lie outside of the SVZ and RMS, whereas processes run along, but rarely cross, the boundaries of the neurogenic BrdUrd+ areas. Brackets indicate areas of lower NOS staining in the SVZ (a and c) and RMS (a and b) as compared with neighboring parenchyma. In all panels the arrows indicate examples of cell bodies of NOS+ neurons, and arrowheads show varicose NOS+ processes that are proximal to dividing cells (c and d). (Insets) Anatomical position of photomicrographs of sagittal rat brain sections. cc, corpus callosum; ctx, cortex; str, striatum. (Scale bars: a, 1 mm; b, 200 μm; c and d, 50 μm.)
These data demonstrate that the areas of active neurogenesis in the adult brain are largely free of, but closely apposed by, NOS-expressing cells. This distribution suggests a link between NO signaling and control of cell division in the neurogenic areas of the brain.
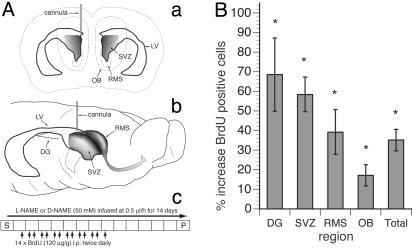
NOS Inhibition Causes Increased Cell Proliferation in Neurogenic Zones of Adult Brain. To address the role of NO during adult neurogenesis, we introduced a specific inhibitor of NOS, l-NAME, into the LV of the adult rat brain using osmotic minipumps (19). This compound is a competitive inhibitor of NOS with slow kinetics for binding and reversal and an ≈10-fold higher affinity for the constitutive NOS isoforms (nNOS and endothelial NOS) over inducible NOS (12). The zone of infusion into the LV was immediately proximal to the anterior SVZ, the major site of adult neurogenesis (Fig. 2A a and b). This infusion of l-NAME or its inactive enantiomer d-NAME was accompanied by i.p. injections of BrdUrd (Fig. 2 Ac). The brains then were sectioned and analyzed for BrdUrd incorporation and for the expression of phenotype-specific markers (5, 11, 20, 21).
Fig. 2.
(A) Scheme of l-NAME or d-NAME infusion and BrdUrd administration. (a and b) Projection diagrams of the relationship of the infusion point into the LV of the rat brain showing anatomy including the SVZ, RMS, LV, OB, and DG. (c) The protocol for administration of BrdUrd and l- or d-NAME in relation to the time for surgery (S), infusion of drugs, and perfusion (P). (B) Changes in the numbers of BrdUrd+ nuclei after persistent NOS inhibition. Absolute counts of BrdUrd+ nuclei of different regions from serially sampled sagittal sections of l-NAME-infused animals show increases in all areas examined when compared with d-NAME-treated animals. The “total” column represents the sum of all the regions examined. The Mann–Whitney test shows that the differences in all tested regions were significant (P < 0.05; see Table 1). DG here refers to the S-GCL and GCL.
We found a robust and significant (P < 0.05, Mann–Whitney test) increase in the number of BrdUrd+ cells in neurogenic regions (Fig. 2B and Table 1: the SVZ (58.4%), RMS (39.2%), and olfactory bulb (OB) (17.1%). A strong increase in the number of BrdUrd+ cells was also evident in the GCL and the S-GCL of the DG (68.5%, P < 0.05). We also saw a significant increase in proliferation in the nonneurogenic regions of the striatum (85.7%, P = 0.005) and the remaining (non-DG) hippocampus (70.3%, P = 0.02). In control experiments we showed that l-NAME was an effective inhibitor for the entire period of the experiment and that prolonged infusion of l-NAME did not affect the distribution of NOS as revealed by NADPH-diaphorase histochemistry or nNOS immunocytochemistry (data not shown). Together our results are consistent with the notion that inhibition of NOS in vivo causes increased proliferation in neurogenic zones of the adult brain.
Table 1. Cells counted in sampled rat brain sections.
| Region | d-NAME | l-NAME | % Increase |
|---|---|---|---|
| DG (S-)GCL | 695 ± 146 | 1,171 ± 225 | 68.5* |
| SVZ | 3,836 ± 772 | 6,076 ± 584 | 58.4* |
| RMS | 11,646 ± 815 | 16,213 ± 2,299 | 39.2* |
| OB | 18,971 ± 934 | 22,223 ± 1,791 | 17.1* |
| All counted regions above together | 38,694 ± 2,517 | 52,269 ± 3,704 | 35.1* |
Average number of BrdUrd+ cells for three brains in each treatment ± the standard deviation. The total number of BrdUrd+ cells counted in all six brains was 272,890
Statistical significance (P < 0.05) using the Mann-Whitney test
To test whether the observed changes were induced in response to damage to the brain tissue invoked by the experimental procedures (4, 22, 23), we looked for changes in the distribution of glial cells among the BrdUrd+ cells. We examined the differences in programmed cell death between brains of animals either treated with inhibitor or control solution or left untreated. We found that there was an increase in the number of BrdUrd+ cells expressing glial fibrillary acidic protein only in the immediate vicinity of the cannula in the brain parenchyma of both l- and d-NAME-treated animals, which is consistent with activated astrocytes being born or migrating to the site of trauma induced by cannulation. However, these changes were limited to the path of the cannula, they were not observed in the areas used to derive the data in Fig. 2B and were not different in animals treated either with NOS inhibitor or its inactive counterpart (data not shown), suggesting that the changes presented in Fig. 2B and Table 1 were not caused by a damage response.
The increase in the number of BrdUrd+ cells after exposure to NOS inhibitor may be a result of increased cell division or decreased cell death. To address this issue we examined the extent of apoptosis in the SVZ and the RMS by using the TUNEL assay. We did not observe differences in the number or distribution of TUNEL-positive cells between the animals exposed to NOS inhibitor and control animals either exposed to inactive enantiomer or left untreated (Kruskal–Wallis test, P < 0.05). The fractions of TUNEL-positive cells in the SVZ and the RMS of l-NAME-treated animals were 1.43% and 0.82%, respectively, and were not significantly different from the percentages obtained with the untreated or d-NAME-treated animals; they were also consistent with the published data on apoptosis in the SVZ and RMS (7). Together, these experiments indicate that the increased number of BrdUrd+ cells seen after NOS inhibition is not a reflection of decreased programmed cell death but is a result of increased cell division.
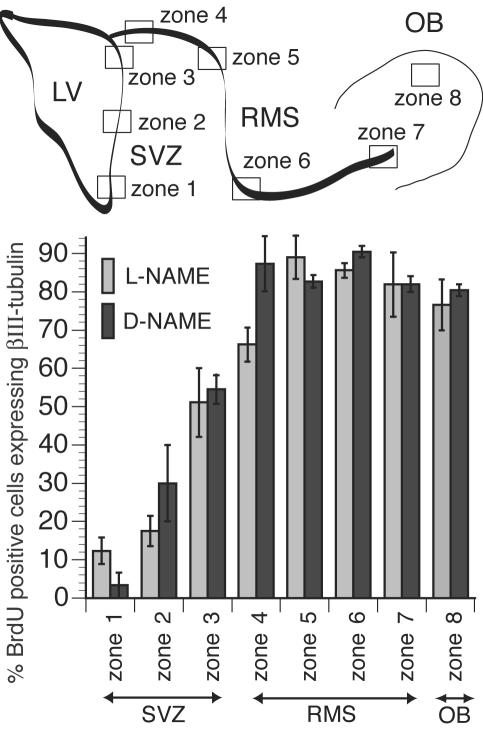
The Phenotypic Fate of New Cells Generated After NOS Inhibition Matches Those Normally Produced. To determine the fate of new cells generated in the adult brain after exposure to NOS inhibitor, we identified the phenotype of newly generated cells in neurogenic regions by using cell-type-specific markers. We stained brain sections with BrdUrd-specific antibody and an antibody against neuron-specific βIII-tubulin, to identify newly generated neurons. We used confocal microscopy to identify βIII-tubulin/BrdUrd double-labeled cells (5) in selected zones along the SVZ and the RMS (Fig. 3); these zones reflect the migration of newly generated neurons from the LV to the OB. In the ventral aspects of the anterior SVZ (zone 1), few BrdUrd+ nuclei (≈10%) also stained for βIII-tubulin. The degree of colocalization increased in more dorsal regions of the anterior SVZ and reached ≈66% in the most proximal region of the RMS (zone 4) and ≈87% in more distal regions (zone 5). The majority of the BrdUrd+ cells (80–90%) along the RMS (zones 5–7) and in the OB (zone 8) also expressed βIII-tubulin, indicating that most of the newly generated cells in these areas are colabeled and undergoing neuronal differentiation. In none of the examined zones were there any significant differences in βIII-tubulin/BrdUrd colocalization between data derived from experimental (L-NAME) and control (D-NAME) animals. These results suggest that introduction of NOS inhibitors increased proliferation of neuronal progenitors without altering the phenotype of the cells being generated.
Fig. 3.
NOS inhibition does not change the fate of cells born in the SVZ and RMS and destined for the OB. A highly schematic diagram of neurogenic zones in adult rodent brains is shown (Upper) to identify the zones for the histogram (Lower). The percentage of cells that have incorporated BrdUrd and express the neuronal marker βIII-tubulin was compared in several arbitrarily defined zones along the SVZ, RMS, and OB for l-NAME-treated and control (d-NAME-treated) animals. Scoring was performed by using confocal z series of images through the sections with orthogonal projections as shown for the data in Fig. 4. There was no statistically significant difference between these treatments for each zone tested, suggesting that the choice of neuronal fate by the majority of new cells in these regions was not affected despite the numbers of new cells being significantly increased in l-NAME-treated animals (Fig. 2B). Mann–Whitney test: P = 0.121, 0.154, 0.827, 0.077, 0.275, 0.105, 0.653, and 0.513 for zones 1–8, respectively.
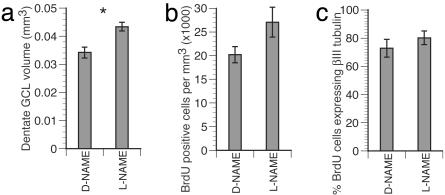
Similarly, in the DG of the hippocampus, NOS inhibition resulted in a significant increase in the number of BrdUrd+ cells in the S-GCL and GCL (Fig. 2B and Table 1), and the fraction of cells labeled with βIII-tubulin among BrdUrd+ cells was unchanged (Figs. 4 and 5c). The increase in the number of BrdUrd+ cells in the DG (Fig. 2B) was accompanied by a significant increase in the volume of the GCL in the sampled sections (27%, P < 0.05, Mann–Whitney test; Fig. 5a) (although not in the density of BrdUrd+ cells; Fig. 5b), suggesting that new cells induced here directly contribute to the cytoarchitecture of the hippocampus. Thus, NOS inhibition stimulated hippocampal neurogenesis in the adult brain. Together, our results demonstrate that suppression of NOS activity augments the generation of new neurons in the adult brain.
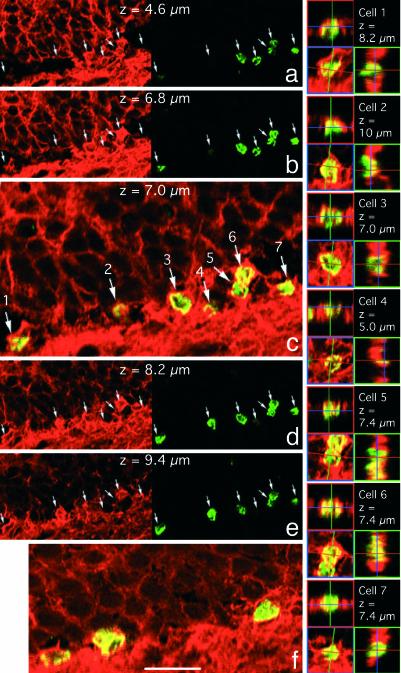
Fig. 4.
New cells in the DG choose neuronal fate. (a–e) Confocal microscope optical sections at 1.2-μm intervals from the DG of an animal exposed to l-NAME. (f) Representative section from the DG of the control d-NAME-treated animal. a, b, d, and e show the βIII-tubulin (red, Left) and BrdUrd (green, Right) staining; c and f show merged channels. Note that the scale on c is twice the scale on a, b, d, and e. Each cell in c is identified with a number, and the orthogonal projection for this cell is presented on the right. In this field of view of seven cells in the DG S-GCL, only cell number 4 does not meet our criteria for coimmunostaining for both BrdUrd and βIII-tubulin as described in Supporting Materials and Methods. The numbers of BrdUrd+ cells expressing βIII-tubulin are quantitated in Fig. 5c. (Scale bars: c, f, and the orthogonal projections, 20 μm; a, b, d, and e, 40 μm.)
Fig. 5.
New cells born after NOS inhibition contribute to the GCL of the DG. The GCL volume increased significantly in l-NAME-treated brain (a, P < 0.05). The change in number of BrdUrd+ cells per volume (b, density) was not significant (P = 0.127), even though there was a 68.5% increase in BrdUrd cell numbers in this region (Fig. 2B) after l-NAME treatment. The proportion of cells expressing BrdUrd and βIII-tubulin was not different between treatments (c, P = 0.564), suggesting that the fate of new cells in the DG was not changed.
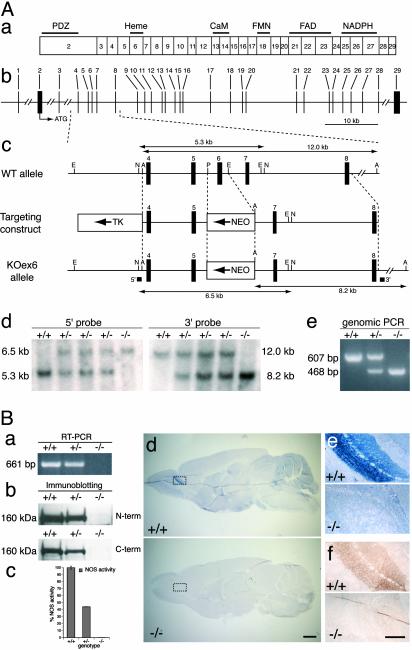
Targeted Disruption of the nNOS Gene Results in an Increase in Proliferation in Neurogenic Zones. To confirm that NO may serve as a negative regulator of cell division in the adult brain, we decided to complement and extend our pharmacological studies with genetic investigations. Because nNOS is the enzyme responsible for producing the majority of NO in the adult mammalian brain (24), we decided to create a loss-of-function (null) mutant of the nNOS gene by targeting the essential heme-binding site of the nNOS enzyme (25, 26); this site is largely encoded by the sixth exon of the mouse nNOS gene (Fig. 6Aa).
Fig. 6.
Targeted inactivation of the mouse nNOS gene. (A) Targeting strategy. (a) ORF of the nNOS gene. Exons are numbered, and regions encoding structural domains and cofactor-binding sites are shown. (b) Structural organization of the nNOS locus (according to GenBank accession no. NT_039312). (c) Structure of WT allele, targeting construct, and targeted KOex6 allele. Thick vertical bars, exons; thin vertical lines, restriction enzymes sites (A, ApaI; E, EcoRI; N, NheI; P, PmlI); arrows, the directions of transcription of the neomycin phosphotransferase gene (NEO) and the thymidine kinase gene (TK); thick horizontal bars, the 3′ and the 5′ hybridization probes used for Southern blot analysis; thin horizontal lines with arrowheads, ApaI and NheI fragments detected with 3′ and 5′ probes, respectively. (d) Southern blot analysis of mouse tail DNA. The 5.3- and 6.5-kb NheI fragments detected with the 5′ probe correspond to the WT allele and to the properly targeted KOex6 allele, respectively. The 12.0- and 8.2-kb ApaI fragments detected with the 3′ probe correspond to the WT allele and to the properly targeted KOex6 allele, respectively. (e) PCR analysis of mouse tail DNA with two pairs of primers for detection of the WT allele (607-bp fragment) and the KOex6 allele (468-bp fragment). (d and e) The sample genotype is indicated above blot or gel. (B) Removal of exon 6 results in complete inactivation of the nNOS gene. (a) RT-PCR analysis of total RNA isolated from whole mouse brains. A pair of primers used detects the 661-bp cDNA product of the WT nNOS allele (+/+,+/-), which is absent in KOex6(-/-) RNA sample. (b) Western blot analysis of whole mouse brain protein extracts. Blots were probed with antibodies specific either to the amino terminus (N-term) or carboxyl terminus (C-term) of the nNOS. A 160-kDa protein band corresponding to the nNOS was not detected in KOex6 (-/-) brain sample. Note that expression levels of nNOS enzyme in WT brains were approximately two times higher than in heterozygous brains. (c) NOS catalytic activity in whole brain protein extracts. Note that enzymatic activity in KOex6 brain homogenates was at background levels (0.15% of that in WT). NOS activity in heterozygous brains was 44.2%, approximately half that of WT, which correlates with expression levels of the nNOS in WT and nNOS+/- brains according to immunoblotting data (Fig. 6Bb). Taken together these results suggest that there is no nNOS allelic compensation in mice heterozygous for the KOex6 mutation. (d–f) The absence of nNOS in the brains of adult KOex6 mice is evident after diaphorase staining of the whole brain (d) and the OB (e) and by immunohistochemistry in the OB with anti-nNOS antibody (f). (e and f) Higher-power images of the regions indicated by the boxes in d. (Scale bars: d, 1 mm; e–f, 100 μm.) In a–f, the sample genotype is indicated.
Mice with a mutated nNOS gene have been generated previously by targeting the second exon of the gene (24) (which does not encode catalytically important regions), and these animals have become valuable tools for studying the role of nNOS. However, the use of alternative promoters and splice sites in these mice results in the expression of a number of nNOS RNA and protein isoforms and concomitant production of NO in the brain by residual nNOS activity (24, 27, 28). To generate a null mutant of nNOS we targeted exon 6 of the gene. We tested several nNOS deletion constructs and found that in-frame removal of exon 6 leads to complete loss of enzymatic activity after transfection into cultured cells (data not shown). Thus, we generated a targeting construct by replacing exon 6 and the adjacent intronic sequences with the neomycin phosphotransferase gene and used it to produce homozygous mutant mice deficient in nNOS activity (KOex6 mice; Fig. 6 Ab–Ae). Full-length nNOS mRNA was not detected in these mice when using RT-PCR amplification (Fig. 6Ba), although low levels of transcripts lacking exon 6 were found in the brain (data not shown). Western blot analysis with antibodies to either the amino- or carboxyl-terminal region of nNOS failed to detect any nNOS protein (Fig. 6Bb). Furthermore, no diaphorase staining (Fig. 6 Bd and Be) nor nNOS immunoreactivity (Fig. 6Bf) was seen in the brain of KOex6 animals. Importantly, we were able to detect only negligible levels of NOS enzymatic activity in brain homogenates of KOex6 mutant mice (0.15 ± 0.11% of the levels seen in WT mice; Fig. 6Bc), in contrast to the animals lacking exon 2, which had a substantial level of residual NOS enzymatic activity in the brain (up to 7% of WT level in some regions) (24). Together, these results demonstrate that we have generated a true null mutant of the nNOS gene with a complete loss of function.
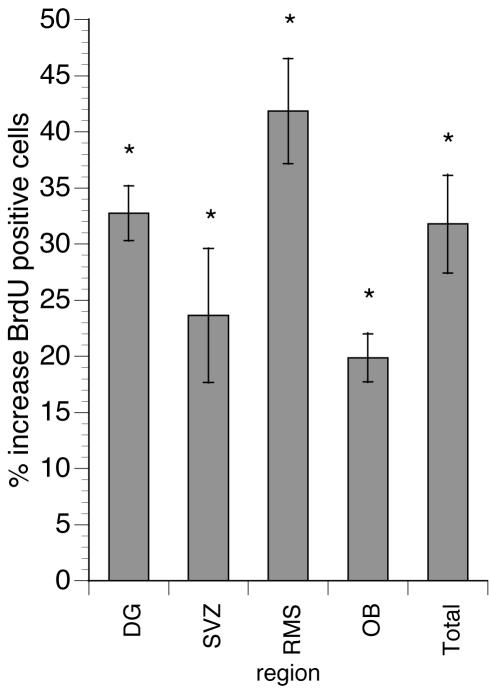
We used these mutant mice to examine the role of NO during adult neurogenesis. We labeled dividing cells with BrdUrd and analyzed sections of adult KOex6 brains for the number and distribution of BrdUrd+ cells. We found a significant (P < 0.05, Mann–Whitney test) increase in the number of BrdUrd-stained nuclei in the neurogenic regions of KOex6 mouse brains as compared with their WT littermates: 23.7% in the SVZ, 41.8% in the RMS, 19.9% in the OB, and 32.7% in the DG (Fig. 7 and Table 2). These results demonstrate that cell division in the neurogenic areas of the adult brain is regulated by the production of NO, paralleling the data obtained when introducing inhibitors of NOS into the rat brain.
Fig. 7.
Increased cell proliferation in various neurogenic regions of the brain in KOex6 adult mice. The absolute numbers of BrdUrd+ cells in different neurogenic regions from serially sampled sagittal sections of KOex6 animals show increases in all areas examined when compared with their WT littermates (Table 2). The “total” column represents the sum of all the regions examined. The Mann–Whitney test shows that the difference in all tested regions was significant (*, P < 0.05).
Table 2. Cells counted in sampled mouse brain sections.
| Region | WT | KOex6 | % Increase |
|---|---|---|---|
| DG | 357 ± 41 | 474 ± 15 | 32.7* |
| SVZ | 2,767 ± 181 | 3,422 ± 286 | 23.7* |
| RMS | 4,467 ± 438 | 6,337 ± 363 | 41.8* |
| OB | 2,188 ± 242 | 2,623 ± 81 | 19.9* |
| All counted regions above together | 9,779 ± 542 | 12,886 ± 739 | 31.8* |
Average number of BrdUrd+ cells for three brains of each genotype (2-month-old females) ± the standard deviation. The total number of BrdUrd+ cells counted in all six brains was 67,995
Statistical significance (P < 0.05) using the Mann-Whitney test
When the distribution of BrdUrd+ cells was examined in the brains of KOex6 mice, the changes in cell proliferation evoked by the mutation were particularly obvious: A strong increase in the number of BrdUrd+ cells was clearly evident in the OB, RMS, and DG. Interestingly, not only did the RMS contain a higher number of dividing cells, but some of these cells appeared to be migrating away from the RMS toward the cortex; many of them appeared grouped into strings that extended into the corpus callosum (data not shown).
Similar results were thus obtained by using two quite distinct experimental paradigms (pharmacological inhibition of NOS activity and genetic inactivation of nNOS activity), which provides strong evidence that NO is a negative regulator of cell proliferation in neurogenic regions of the adult mammalian brain.
Discussion
Here we report that inhibition of NOS activity augments neurogenesis in the neurogenic areas of the adult brain. These changes are due to an increase in cell division and not a decrease in programmed cell death. Specifically, a reduction of NOS activity stimulates production of new neurons in both the forebrain subependyma and hippocampal DG, two regions that generate neurons constitutively throughout adulthood. In these regions several subpopulations of neural stem and progenitor cells have been characterized. In particular, in the ventricular subependyma, subpopulations of dividing neuronal progenitors have been identified in addition to more quiescent cells with astrocyte-like properties (29). NO signaling may help to maintain the quiescence of neural stem cells or to promote terminal differentiation in advanced neuronal progenitors; NO may also exert its antiproliferative activity at multiple points in the neuronal differentiation cascade.
NO may be produced in an autocrine fashion by neural stem or progenitor cells or in a paracrine fashion by surrounding cells. Our observations that NOS+ neurons are largely excluded from the neurogenic areas of the adult rat brain confirm previous findings in mouse (30, 31) and are consistent with the notion of paracrine signaling. NO may be produced in the layers adjacent to the dividing cells or may be released from the processes of nitrergic neurons within the SVZ and the RMS. The high density of nNOS+ cells in nonneurogenic areas such as striatum raises the possibility that NOS may chronically suppress a dormant reserve of progenitor cells in such areas of the brain.
In our experiments we delivered NOS inhibitor to the ventricular areas by using continuos intraventricular infusion. Less-direct approaches to manipulate NO signaling in the organism may affect multiple physiological responses, particularly when used in a confounding context of organ injury; for instance, i.p. introduction of NO donors or inhibitors on the background of ischemia affects neurogenesis, perhaps by activating antiapoptotic, vascular, or systemic responses (32, 33).
The ability of NO to suppress cell division has been demonstrated in neural development in a number of settings. NO acts as a negative regulator of cell proliferation in the optic tectum of the Xenopus tadpole (34). NO synthesis is required for neurogenesis in the optic lobe of moth (35) as well as for neuronal differentiation of cells in culture (36–38). NO also modulates the balance between cell proliferation and cell differentiation in systems as diverse as developing Drosophila imaginal disks and embryos (16, 39, 40) and numerous types of cultured cells (for review see ref. 16).
The molecular bases for the antiproliferative action of NO are only beginning to be understood. Evidence from several experimental models suggest that p21 may mediate some of the NO action in G1 phase (37, 41). Genetic data indicate that NO interacts with the retinoblastoma (Rb) pathway to control cell division in Drosophila, and we have suggested that Rb protein may be a direct target for S-nitrosylation (42). Interestingly, Rb is one of several proteins that were found in a nitrosylated state in a search for S-nitrosylated proteins (43). Yet another plausible target is ornithine decarboxylase, since its S-nitrosylation may inhibit proliferation of smooth muscle cells (41). Furthermore, our recent data indicate that NO affects multiple checkpoints in the cell cycle (N. Nakaya and G.E., unpublished data).
Several lines of evidence have suggested negative regulation of adult neurogenesis (44–48), although the signals that suppress the birth of new neurons in the adult brain remain unclear. In our study we used two independent lines of evidence to identify NO as one such suppressor of adult neurogenesis. Our pharmacological experiments with NOS inhibitors implicate NO as an important suppressive signal, and the loss-of-function genetic model confirmed that nNOS was indeed the target of our pharmacological intervention. These findings strongly support the role of NO as an essential negative regulator of cell proliferation in the adult mammalian brain. Furthermore, they indicate that adult neurogenesis may be augmented by therapeutic intervention aimed at a clearly defined molecular target.
Supplementary Material
Acknowledgments
We thank Kim Lerner and John Mignone for help with related experiments; Isabel Cantallops for advice on statistical analysis; Enikolopov and Goldman Laboratory members for useful discussion; and Julian Banerji for critical reading of the manuscript. This work was funded in the Enikolopov Laboratory by the National Institutes of Health, the Seraph Foundation, the Charles Henry Leach II Foundation, and the Donaldson Foundation (G.E.); the New Zealand Foundation for Research, Science, and Technology (M.A.P.); and in the Goldman Laboratory by National Institute of Neurological Disorders and Stroke, Project ALS, the Christopher Reeve Paralysis Foundation, the National Multiple Sclerosis Society, and the Cure HD initiative of the Hereditary Disease Foundation.
Abbreviations: SVZ, subventricular zone; LV, lateral ventricle; RMS, rostral migratory stream; GCL, granular cell layer; S-GCL, sub-GCL; DG, dentate gyrus; NOS, NO synthase; nNOS, neuronal NOS; NAME, Nω-nitro-methyl-L[D]-arginine methyl ester; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP end labeling; KOex6, disrupted nNOS allele; OB, olfactory bulb.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF534819, AF534820, and AF534821).
References
- 1.Gage, F. H. (2000) Science 287, 1433-1438. [DOI] [PubMed] [Google Scholar]
- 2.Goldman, S. A. & Luskin, M. B. (1998) Trends. Neurosci. 21, 107-114. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Buylla, A., Garcia-Verdugo, J. M. & Tramontin, A. D. (2001) Nat. Rev. Neurosci. 2, 287-293. [DOI] [PubMed] [Google Scholar]
- 4.Magavi, S. S., Leavitt, B. R. & Macklis, J. D. (2000) Nature 405, 951-955. [DOI] [PubMed] [Google Scholar]
- 5.Benraiss, A., Chmielnicki, E., Lerner, K., Roh, D. & Goldman, S. A. (2001) J. Neurosci. 21, 6718-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pencea, V., Bingaman, K. D., Wiegand, S. J. & Luskin, M. B. (2001) J. Neurosci. 21, 6706-6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biebl, M., Cooper, C. M., Winkler, J. & Kuhn, H. G. (2000) Neurosci. Lett. 291, 17-20. [DOI] [PubMed] [Google Scholar]
- 8.Song, H. J., Stevens, C. F. & Gage, F. H. (2002) Nat. Neurosci. 5, 438-445. [DOI] [PubMed] [Google Scholar]
- 9.Craig, C. G., Tropepe, V., Morshead, C. M., Reynolds, B. A., Weiss, S. & van der Kooy, D. (1996) J. Neurosci. 16, 2649-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aberg, M. A., Aberg, N. D., Hedbacker, H., Oscarsson, J. & Eriksson, P. S. (2000) J. Neurosci. 20, 2896-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn, H. G., Winkler, J., Kempermann, G., Thal, L. J. & Gage, F. H. (1997) J. Neurosci. 17, 5820-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ignarro, L. J. (2000) Nitric Oxide: Biology and Pathobiology (Academic, San Diego).
- 13.Bredt, D. S. & Snyder, S. H. (1994) Neuron 13, 301-313. [DOI] [PubMed] [Google Scholar]
- 14.Garg, U. C. & Hassid, A. (1990) Biochem. Biophys. Res. Commun. 171, 474-479. [DOI] [PubMed] [Google Scholar]
- 15.Lepoivre, M., Chenais, B., Yapo, A., Lemaire, G., Thelander, L. & Tenu, J. P. (1990) J. Biol. Chem. 265, 14143-14149. [PubMed] [Google Scholar]
- 16.Enikolopov, G., Banerji, J. & Kuzin, B. (1999) Cell Death Differ. 6, 956-963. [DOI] [PubMed] [Google Scholar]
- 17.Dawson, T. M., Bredt, D. S., Fotuhi, M., Hwang, P. M. & Snyder, S. H. (1991) Proc. Natl. Acad. Sci. USA 88, 7797-7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigo, J., Springall, D. R., Uttenthal, O., Bentura, M. L., Abadia-Molina, F., Riveros-Moreno, V., Martinez-Murillo, R., Polak, J. M. & Moncada, S. (1994) Philos. Trans. R. Soc. London B 345, 175-221. [DOI] [PubMed] [Google Scholar]
- 19.Paxinos, G. & Watson, C. (1998) The Rat Brain in Stereotactic Coordinates (Academic, San Diego).
- 20.Coggeshall, R. E. & Lekan, H. A. (1996) J. Comp. Neurol. 364, 6-15. [DOI] [PubMed] [Google Scholar]
- 21.Guillery, R. W. & Herrup, K. (1997) J. Comp. Neurol. 386, 2-7. [DOI] [PubMed] [Google Scholar]
- 22.Weinstein, D. E., Burrola, P. & Kilpatrick, T. J. (1996) Brain Res. 743, 11-16. [DOI] [PubMed] [Google Scholar]
- 23.Jin, K., Minami, M., Lan, J. Q., Mao, X. O., Batteur, S., Simon, R. P. & Greenberg, D. A. (2001) Proc. Natl. Acad. Sci. USA 98, 4710-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, P. L., Dawson, T. M., Bredt, D. S., Snyder, S. H. & Fishman, M. C. (1993) Cell 75, 1273-1286. [DOI] [PubMed] [Google Scholar]
- 25.Klatt, P., Pfeiffer, S., List, B. M., Lehner, D., Glatter, O., Bachinger, H. P., Werner, E. R., Schmidt, K. & Mayer, B. (1996) J. Biol. Chem. 271, 7336-7342. [DOI] [PubMed] [Google Scholar]
- 26.Richards, M. K., Clague, M. J. & Marletta, M. A. (1996) Biochemistry 35, 7772-7780. [DOI] [PubMed] [Google Scholar]
- 27.Eliasson, M. J., Blackshaw, S., Schell, M. J. & Snyder, S. H. (1997) Proc. Natl. Acad. Sci. USA 94, 3396-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brenman, J. E., Xia, H., Chao, D. S., Black, S. M. & Bredt, D. S. (1997) Dev. Neurosci. 19, 224-231. [DOI] [PubMed] [Google Scholar]
- 29.Doetsch, F., García-Verdugo, J. M. & Alvarez-Buylla, A. (1997) J. Neurosci. 17, 5046-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gates, M. A., Thomas, L. B., Howard, E. M., Laywell, E. D., Sajin, B., Faissner, A., Gotz, B., Silver, J. & Steindler, D. A. (1995) J. Comp. Neurol. 361, 249-266. [DOI] [PubMed] [Google Scholar]
- 31.Moreno-Lopez, B., Noval, J. A., Gonzalez-Bonet, L. G. & Estrada, C. (2000) Brain Res. 869, 244-250. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, R., Zhang, L., Zhang, Z., Wang, Y., Lu, M., Lapointe, M. & Chopp, M. (2001) Ann. Neurol. 50, 602-611. [DOI] [PubMed] [Google Scholar]
- 33.Zhu, D. Y., Liu, S. H., Sun, H. S. & Lu, Y. M. (2003) J. Neurosci. 23, 223-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peunova, N., Scheinker, V., Cline, H. & Enikolopov, G. (2001) J. Neurosci. 21, 8809-8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Champlin, D. T. & Truman, J. W. (2000) Development (Cambridge, U.K.) 127, 3543-3551. [DOI] [PubMed] [Google Scholar]
- 36.Peunova, N. & Enikolopov, G. (1995) Nature 375, 68-73. [DOI] [PubMed] [Google Scholar]
- 37.Poluha, W., Schonhoff, C. M., Harrington, K. S., Lachyankar, M. B., Crosbie, N. E., Bulseco, D. A. & Ross, A. H. (1997) J. Biol. Chem. 272, 24002-24007. [DOI] [PubMed] [Google Scholar]
- 38.Côté, F., Laflamme, L., Payet, M. D. & Gallo-Payet, N. (1998) Endocr. Res. 24, 403-407. [DOI] [PubMed] [Google Scholar]
- 39.Kuzin, B., Roberts, I., Peunova, N. & Enikolopov, G. (1996) Cell 87, 639-649. [DOI] [PubMed] [Google Scholar]
- 40.Wingrove, J. A. & O'Farrell, P. H. (1999) Cell 98, 105-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauer, P. M., Buga, G. M., Fukuto, J. M., Pegg, A. E. & Ignarro, L. J. (2001) J. Biol. Chem. 276, 34458-34464. [DOI] [PubMed] [Google Scholar]
- 42.Kuzin, B., Regulski, M., Stasiv, Y., Scheinker, V., Tully, T. & Enikolopov, G. (2000) Curr. Biol. 10, 459-462. [DOI] [PubMed] [Google Scholar]
- 43.Jaffrey, S. R., Erdjument-Bromage, H., Ferris, C. D., Tempst, P. & Snyder, S. H. (2001) Nat. Cell Biol. 3, 193-197. [DOI] [PubMed] [Google Scholar]
- 44.Morshead, C. M. & van der Kooy, D. (1992) J. Neurosci. 12, 249-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldman, S. A., Zaremba, A. & Niedzwiecki, D. (1992) J. Neurosci. 12, 2532-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morshead, C. M., Reynolds, B. A., Craig, C. G., McBurney, M. W., Staines, W. A., Morassutti, D., Weiss, S. & van der Kooy, D. (1994) Neuron 13, 1071-1082. [DOI] [PubMed] [Google Scholar]
- 47.Kirschenbaum, B. & Goldman, S. A. (1995) Proc. Natl. Acad. Sci. USA 92, 210-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palmer, T. D., Takahashi, J. & Gage, F. H. (1997) Mol. Cell. Neurosci. 8, 389-404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.