Abstract
Multistep proteolytic mechanisms are essential for converting proprotein precursors into active peptide neurotransmitters and hormones. Cysteine proteases have been implicated in the processing of proenkephalin and other neuropeptide precursors. Although the papain family of cysteine proteases has been considered the primary proteases of the lysosomal degradation pathway, more recent studies indicate that functions of these enzymes are linked to specific biological processes. However, few protein substrates have been described for members of this family. We show here that secretory vesicle cathepsin L is the responsible cysteine protease of chromaffin granules for converting proenkephalin to the active enkephalin peptide neurotransmitter. The cysteine protease activity was identified as cathepsin L by affinity labeling with an activity-based probe for cysteine proteases followed by mass spectrometry for peptide sequencing. Production of [Met]enkephalin by cathepsin L occurred by proteolytic processing at dibasic and monobasic prohormone-processing sites. Cellular studies showed the colocalization of cathepsin L with [Met]enkephalin in secretory vesicles of neuroendocrine chromaffin cells by immunofluorescent confocal and immunoelectron microscopy. Functional localization of cathepsin L to the regulated secretory pathway was demonstrated by its cosecretion with [Met]enkephalin. Finally, in cathepsin L gene knockout mice, [Met]enkephalin levels in brain were reduced significantly; this occurred with an increase in the relative amounts of enkephalin precursor. These findings indicate a previously uncharacterized biological role for secretory vesicle cathepsin L in the production of [Met]enkephalin, an endogenous peptide neurotransmitter.
The biosynthesis of enkephalin opioid peptides as well as numerous peptide neurotransmitters and hormones requires proteolytic processing of respective proprotein precursors within regulated secretory vesicles (1–4). The mature, processed enkephalin peptide is stored within these vesicles and undergoes stimulated secretion to mediate neurotransmission and cell–cell communication in the regulation of analgesia, behavior, and immune-cell functions. Secretory vesicles of neuroendocrine chromaffin cells (also known as chromaffin granules) contain enkephalin and its precursor proenkephalin (PE) (5, 6), with relevant prohormone convertases for converting PE into active enkephalin.
The primary PE-cleaving activity in chromaffin granules has been characterized as a cysteine protease complex known as “prohormone thiol protease” (PTP) (7–10). The cysteine protease activity cleaves PE and enkephalin-containing peptide substrates at paired basic residues, as well as at certain monobasic residues, to generate appropriate enkephalin-related peptide products. Cellular inhibition of PTP by a cysteine protease inhibitor results in reduced production of enkephalin (11). Molecular identification of the protease component responsible for this cysteine protease activity will facilitate our understanding of multiple proteolytic enzymes that produce active peptides including the opioid [Met]enkephalin (ME) (12, 13).
In this study the protease responsible for PE-cleaving activity in chromaffin granules was identified by using an activity-based probe for cysteine proteases (14, 15) combined with mass spectrometry (MS) for peptide sequencing. Results identified secretory vesicle cathepsin L as the enzyme responsible for the previously described PTP cysteine protease activity involved in enkephalin and neuropeptide production (7–10). Cathepsin L generated the active peptide ME by cleaving enkephalin-containing peptide substrates at native dibasic and monobasic sites. Notably, cathepsin L colocalized with ME in the regulated secretory pathway of chromaffin cells. In cathepsin L gene knockout (KO) mice (16–18), ME levels in brain were reduced significantly; an increase in relative amounts of enkephalin precursor also occurred. These results provide evidence for a previously uncharacterized role for cathepsin L in secretory vesicles for the production of an endogenous peptide neurotransmitter, ME.
Methods
Affinity Labeling of PE-Cleaving Activity with DCG-04, an Activity-Based Probe for Cysteine Proteases. PE-cleaving activity was purified from bovine chromaffin granules as described (10). Fractions enriched for PE-cleaving activity were affinity-labeled with DCG-04 or [125I]DCG-04 (14, 15) and subjected to 1D or 2D SDS/PAGE gels, transferred to nitrocellulose membranes, and detected with streptavidin-horseradish peroxidase. Proteins were observed by silver staining (10).
Peptide Sequencing of the Affinity-Labeled Enzyme by MS. DCG-04-labeled proteins on 2D gels were excised, subjected to “in-gel” digestion with trypsin (http://donatello.ucsf.edu/ingel.html), desalted, and subjected to electrospray mass measurement and low-energy collision-induced dissociation (CID) analyses by using nanospray sample introduction on a QSTAR (MDS Sciex, Concord, ON, Canada) quadrupole orthogonal acceleration/time-of-flight (TOF) hybrid tandem mass spectrometer equipped with a Protona source (Protana, Odense, Denmark). Multiply charged ions detected were selected for subsequent low-energy CID analyses. Peptide masses were analyzed by the Protein Prospector MS-Fit search of the NCBI protein database, and the CID data were subjected to MS-Tag searches (http://prospector.ucsf.edu) or manually interpreted.
Cathepsin L Processing of Enkephalin-Containing Peptides. Bovine adrenal medulla peptide 22 (BAM-22P) and peptide F (100 μM, Peninsula Laboratories) were incubated with human cathepsin L (0.5 ng/50 μl, Athens Research & Technology, Athens, GA) at 30°C for 30 min. Peptide products were analyzed by the Voyager DE-STR matrix-assisted laser desorption ionization (MALDI)-TOF and Pulsar QSTAR mass spectrometers. Observed masses of peptides were compared with theoretical peptide products by using the program PAWS (http://prowl.rockefeller.edu/software/contents.htm).
Colocalization of Cathepsin L with ME in Chromaffin Cells by Confocal Microscopy. Immunofluorescence confocal microscopy of chromaffin cells, performed as described (19), used rabbit anti-cathepsin L (Athens Research & Technology) and mouse anti-ME (Chemicon). Cathepsin L was detected with anti-rabbit IgG-Alexa Fluor 488 (green fluorescence, Molecular Probes), and ME was detected with anti-mouse IgG-Alexa Fluor 594 (red fluorescence). Colocalization of cathepsin L and ME was observed by merged images with the Nikon Eclipse 800 microscope coupled to a PCM-2000 confocal system and analyses with SIMPLE PCI software. Nuclei were observed by 4′,6-diamidino-2-phenylindole staining.
Immunoelectron Microscopy of Cathepsin L and ME in Secretory Vesicles. Secretory vesicles were isolated from bovine adrenal medulla by differential sucrose density centrifugation (10) and fixed in 0.2% glutaraldehyde/2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.2. Samples were osmicated in 2% osmium tetroxide in 0.1 M cacodylate and embedded in Epon 812. Ultrathin sections were partially deosmicated through 1% periodic acid/9% sodium periodate and incubated in 3% normal goat serum in 1× Tris-buffered saline. Sections were incubated with mouse anti-ME (Chemicon) and detected with secondary goat anti-mouse IgG conjugated to 6-nm colloidal gold (Aurion, Wageningen, The Netherlands). Sections were postfixed and immunostained with rabbit anti-cathepsin L (Athens Research & Technology) detected by goat anti-rabbit IgG conjugated to 15-nm colloidal gold. Sections were examined in a Tecnai-12 transmission electron microscope by using a charge-coupled device camera and DIGITAL MICROGRAPH software (Gatan, Pleasanton, CA)
Stimulated Cosecretion of Cathepsin L and ME from Chromaffin Cells. Chromaffin cells in primary culture (2 × 106 cells per well) were stimulated with nicotine (10 μM) or KCl (50 mM) for 15 min, and ME in the secretion medium was measured by an RIA as described (19). For secretion of [35S]cathepsin L, proteins were labeled with [35S]methioninefor4hfollowed by incubation of cells with nicotine (10 μM) or KCl (50 mM) for 15 min as described (20). Secreted [35S]cathepsin L was detected by immunoprecipitation with anti-cathepsin L and SDS/PAGE.
ME in Brains of Cathepsin L Gene KO Mice. Cathepsin L-deficient mice were generated by gene targeting in mouse embryonic stem cells as described (16). Genotyping established WT (+/+) and cathepsin L-deficient (-/-) mice. Brain tissues from mice were homogenized in 1 M acetic acid, heated at 95°C for 10 min, centrifuged (15,000 × g for 15 min), and the supernatant was analyzed for ME by an RIA (19). The RIA did not crossreact with PE, expressed and purified from Escherichia coli as described (10); in addition, the RIA does not crossreact with basic residue extended forms of ME (Arg-ME-Lys, Lys-Arg-ME, ME-Lys, and ME-Lys-Arg). To measure relative amounts of enkephalin precursor compared with ME, ME was measured by an RIA in samples treated with trypsin and carboxypeptidase B (incubated with 1 μg of trypsin for 4 h, followed by 1 h with 1 μg of carboxypeptidase B at 37°C) and in untreated control samples. Protein content was measured with the Bradford reagent (Bio-Rad).
Results
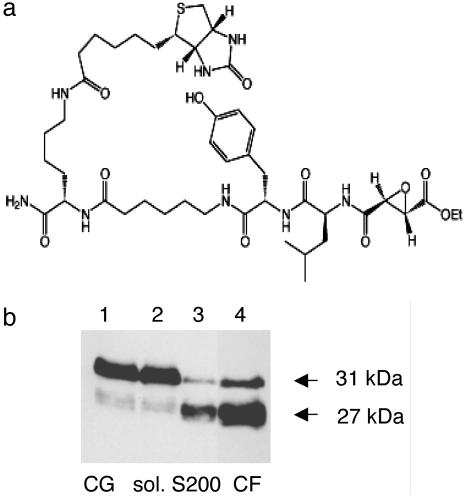
Affinity Labeling of PE-Processing Activity (PTP) in Chromaffin Granules. The PE-processing activity known as PTP exists as a high molecular mass cysteine protease complex (180–200 kDa) that is potently inhibited by the cysteine protease inhibitor E64-c (7–11). To determine the protease component of this activity, affinity labeling was performed with the cysteine protease activity probe known as DCG-04 (Fig. 1a) (14, 15). Affinity labeling at various stages in the purification demonstrated enrichment of a DCG-04-labeled 27-kDa protease band (Fig. 1b) as well as labeling of a less abundant protease of 31 kDa (from S200 gel filtration and chromatofocusing steps; Fig. 1b).
Fig. 1.
DCG-04 affinity labeling of the cysteine protease (PTP) from secretory vesicles. (a) Structure of DCG-04, an activity-based probe for cysteine proteases. The modified cysteine protease inhibitor DCG-04, resulting from biotinylation of E64-c, was used for affinity labeling. (b) Enrichment of the 27-kDa component by DCG-04 affinity labeling during purification of PE-cleaving activity. Relative PE-cleaving activities (PTP) in enriched fractions during purification were 1, 1, 26, and 90 relative units of activity at the steps of lysed chromaffin granules (CG), soluble CG extract (sol.), S200 gel filtration (S200), and chromatofocusing (lanes 1–4, respectively). Protein contents in lanes 1–4 were 170, 110, 2.5, and 1 μg, respectively.
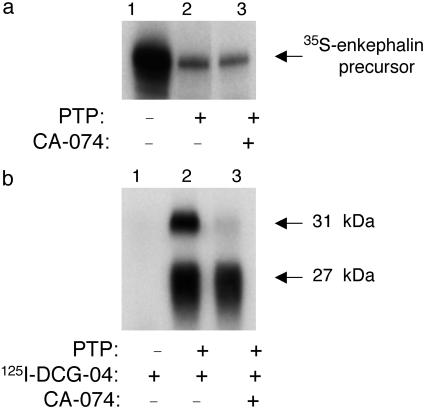
Inhibitor and differential affinity-labeling studies were performed to define the role of the 27- and 31-kDa polypeptides for PE-processing activity. The apparent molecular mass of the 31-kDa protein parallels that of the cysteine protease cathepsin B (21, 22). Therefore, the effect of the selective cathepsin B inhibitor CA-074 (23, 24) on processing activity was tested. CA-074 had no effect on [35S]enkephalin precursor-processing activity (Fig. 2a). Notably, CA-074 did not block DCG-04 labeling of the 27-kDa band (Fig. 2b) but completely blocked the labeling of the 31-kDa polypeptide. These findings indicated that the 27-kDa protein was responsible for PE-processing activity. Moreover, CA-074 competition for DCG-04 labeling of the 31-kDa polypeptide suggested the identity of this protein as cathepsin B.
Fig. 2.
Specific affinity labeling with DCG-04, in the presence of CA-074, identifies the 27-kDa band as the enzyme for PE-cleaving activity. (a) CA-074 does not inhibit PE-cleaving activity. [35S]Enkephalin precursor was incubated with purified PTP in the absence (lane 2) or presence (lane 3) of CA-074 (1 μM) at 37°C for 2 h. Control [35S]enkephalin precursor incubated alone is shown (lane 1). Assays were assessed by SDS/PAGE and autoradiography. (b) DCG-04 affinity labeling of the 27-kDa component is not affected by CA-074. Affinity labeling of PTP by [125I]DCG-04 was conducted in the absence (lane 2) or presence of CA-074 (1 μM, lane 3). Control [125I]DCG-04 alone (no enzyme) was included (lane 1).
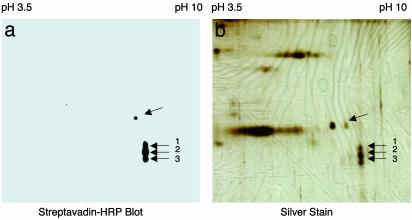
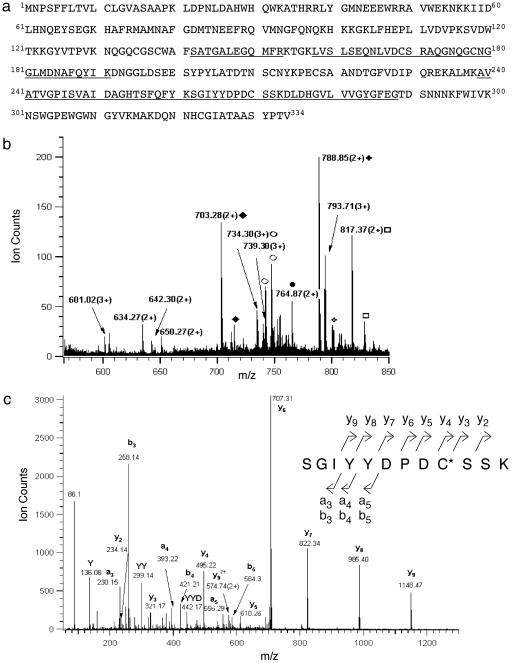
Peptide Sequencing by MS Identifies Cathepsin L as the Cysteine Protease for PE Processing. 2D gels showed that the DCG-04-labeled 27-kDa protein consisted of three spots of similar molecular mass with virtually identical isoelectric points (Fig. 3a). These spots were visualized by silver staining (Fig. 3b), excised, and subjected to tryptic digestion followed by nanoelectrospray MS analyses. Identical tryptic peptide masses and CID (tandem MS) spectra were observed for all three spots, which corresponded to bovine cathepsin L (Fig. 4a). The profile of the 10 tryptic peptides detected by nanoelectrospray MS analyses from spot 2 (Fig. 4b)aswellasfrom spots 1 and 3 corresponded to peptide sequences of cathepsin L represented by residues 142–153, 158–171, 172–191, 239–261, 262–273, and 274–288. In addition, the CID spectrum for the tryptic peptide (residues 262–273) (Fig. 4c) established serine as residue 272, which clarifies earlier sequences submitted to GenBank that indicated cysteine or serine at this position (accession nos. 1705638, 1542853, and 3641698).
Fig. 3.
2D gel analyses of DCG-04 affinity-labeled PE-processing activity. (a) DCG-04-labeled PTP. DCG-04 labeling of purified PE-processing activity on 2D gels was blotted and visualized by avidin-horseradish peroxidase (HRP). (b) DCG-04-labeled enzymes detected on 2D gels. DCG-04-labeled enzyme proteins were subjected to 2D gels and silver staining.
Fig. 4.
Bovine cathepsin L identified by MS. (a) Primary sequence of bovine cathepsin L. Peptides derived from tryptic digests of DCG-04 affinity-labeled 27-kDa proteins, sequenced by CID (tandem MS) MS, are illustrated as the underlined amino acid sequences of bovine cathepsin L. (b) Electrospray mass spectrum of the unfractionated tryptic digest of DCG-04 affinity-labeled 27-kDa enzyme. The electrospray mass spectrum of tryptic peptides of spot 2 from the 2D gel (from Fig. 3a) is illustrated, which were also observed from spots 1 and 3. Primary sequences of these peptides corresponded to bovine cathepsin L, underlined in a. Ions representing the same species (Na-, K- adducts included) are labeled with the same symbol. (c) Low-energy CID spectrum of tryptic peptide (residues 262–273). The tryptic peptide (residues 262–273) was observed from DCG-04-labeled spots 1–3. The CID spectrum for this precursor ion of m/z 703.27(2+) [nomenclature of Biemann (32)] indicated the peptide sequence Ser262-Gly-Ile-Tyr-Tyr-Asp-Pro-Asp-Cys(acrylamide)-Ser-Ser-Lys273, thus establishing Ser-272 instead of Cys-272, which have both been reported in GenBank. C*, acrylamide-modified cysteine.
The three spots of cathepsin L indicate heterogeneity in the molecular forms of secretory vesicle cathepsin L. The identified tryptic peptides and the 26- to 28-kDa apparent molecular mass of the three spots correspond to the single-chain domain of cathepsin L. Because all three spots possess the COOH-terminal tryptic peptide (residues 273–288) of the single-chain form, it is likely that heterogeneity exists in their NH2 termini. In addition, cathepsin L is known to exist as a glycoprotein with glycosylation at Asn-222 (21). Although tryptic peptides containing Asn-222 were not detected in this study, evidence for the glycoprotein nature of the secretory vesicle cathepsin L was suggested by Con A-Sepharose as one of the steps for purification of the PE-cleaving activity (10).
In addition, sequencing by an ion-trap mass spectrometer by microcapillary liquid chromatography tandem MS of tryptic peptides from the 31-kDa spot identified it as bovine cathepsin B (data not shown), thus confirming its predicted identity based on CA-074 inhibition of DCG-04 labeling of this band.
Cathepsin L Cleaves Enkephalin-Containing Peptides at Dibasic and Monobasic Prohormone-Processing Sites. Production of ME by cathepsin L was assessed with the enkephalin-containing peptide substrates BAM-22P and peptide F, with identification of peptide products by MALDI-TOF MS. Cathepsin L, similar to the native cysteine protease activity (PTP), generated active ME by cleaving BAM-22P at the dibasic ↓Arg-↓Arg and monobasic ↓Arg sites (Fig. 5a). Peptide F was cleaved by cathepsin L at dibasic ↓Lys-↓Lys and ↓Lys-Arg sites (Fig. 5b). Some cleavage of peptide F occurred between Gly-Gly within ME (YGGFM), which has been observed for native PTP. Moreover, cathepsin L processing of full-length [35S]enkephalin precursor produced identical products as those generated by native PTP (data not shown). The cleavage specificities of cathepsin L for dibasic and monobasic processing sites are appropriate for generating ME.
Fig. 5.
Cathepsin L cleaves dibasic and monobasic processing sites of enkephalin-containing peptide substrates, BAM-22P, and Peptide F. BAM-22P (a) and Peptide F (b) were incubated with cathepsin L, and peptide products were identified by MALDI-TOF MS. The sequence of ME, YGGFM, is underlined. MS spectra and monoisotopic masses of peptide products are shown in Fig.8, which is published as supporting information on the PNAS web site, www.pnas.org. Cathepsin L cleavage sites are illustrated by solid arrows. Dotted-line arrows indicate secondary cleavage sites.
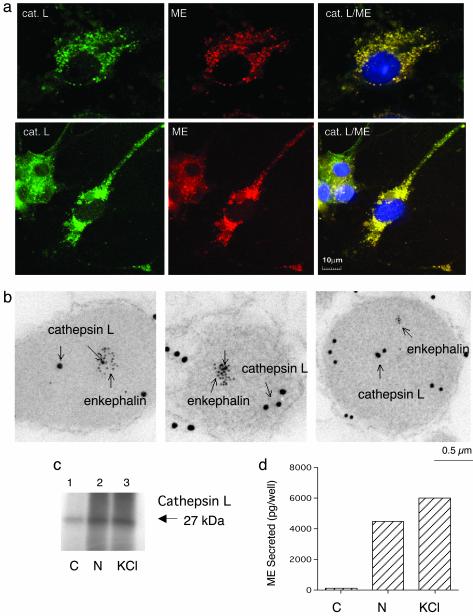
Cellular Colocalization of Cathepsin L with ME in Secretory Vesicles of Neuroendocrine Chromaffin Cells. Involvement of cathepsin L in enkephalin production requires its localization to secretory vesicles that contain PE and ME. Colocalization of cathepsin L with ME was evaluated in these cells by immunofluorescence confocal microscopy (Fig. 6a), combined with immunoelectron microscopy (Fig. 6b). Cathepsin L and ME immunofluorescence were colocalized and appeared as discrete, punctate immunostaining, which is consistent with a secretory vesicle localization. Moreover, neuritic extensions of chromaffin cells displayed discrete colocalization of cathepsin L and ME. Immunoelectron microscopy of cathepsin L and ME in secretory vesicles from chromaffin cells further confirmed their colocalization (Fig. 6b). These results demonstrate the presence of cathepsin L with ME in secretory vesicles.
Fig. 6.
Localization of cathepsin L within enkephalin-containing secretory vesicles of neuroendocrine chromaffin cells. (a) Cathepsin L colocalization with ME by immunofluorescence confocal microscopy. Immunofluorescence localization of cathepsin L (cat. L) was assessed by anti-cathepsin L detected with anti-rabbit IgG-Alexa Fluor 488 (green fluorescence), and ME was detected with anti-ME and anti-mouse IgG-Alexa Fluor 594 (red). Colocalization is illustrated by overlay of the images, which is illustrated by yellow fluorescence. (b) Immunoelectron microscopy demonstrates colocalization of cathepsin L and ME in secretory vesicles. Cathepsin L in secretory vesicles was indicated by anti-cathepsin L detected with 15-nm colloidal gold-conjugated anti-rabbit IgG, and ME was detected with anti-ME and 6-nm colloidal gold-conjugated anti-mouse IgG. The presence of both 15- and 6-nm gold particles within these vesicles demonstrated the in vivo colocalization of cathepsin L and ME. (c) Stimulated secretion of cathepsin L from chromaffin cells. Secretion was induced with nicotine (10 μM) or KCl (50 mM) for 15 min, and [35S]cathepsin L in the secretion medium (with prior [35S]Met labeling) was detected by immunoprecipitation with anti-cathepsin L and SDS/PAGE. Secretion of [35S]cathepsin L was observed in nicotine-(N) and KCl-stimulated cells (lanes 2 and 3, respectively) but not in unstimulated control cells (C) (lane 1). (d) Stimulated secretion of ME from chromaffin cells. ME in the secretion medium from nicotine- or KCl-stimulated cells (for 15 min) (lanes 2 and 3, respectively) and unstimulated control cells (lane 1) were measured by an RIA. Values are averages of triplicate wells ± SEM.
The presence of cathepsin L in enkephalin-containing secretory vesicles predicted its functional cosecretion with ME from chromaffin cells. Therefore, the cosecretion of [35S]cathepsin L and ME during nicotine and KCl stimulation of these cells was evaluated. [35S]Cathepsin L in secretion medium was detected by immunoprecipitation with an anti-cathepsin L antibody, and ME was measured by an RIA. Stimulated secretion of the 27-kDa [35S]cathepsin L occurred with ME during nicotine- and KCl-induced secretion (Fig. 6 c and d, respectively). These findings indicate the presence of cathepsin L in the regulated secretory pathway.
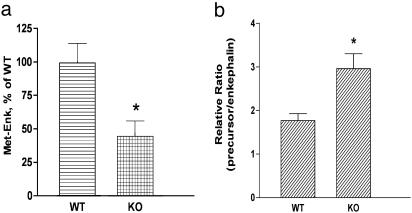
Cathepsin L Gene KO Mice Possess Reduced Levels of ME in Brain. The role of cathepsin L in the production of enkephalin peptides was assessed further in cathepsin L-deficient mice (-/- genotype) compared with WT controls (+/+ genotype). The content of ME in brain was measured by an RIA, which specifically detected ME and not PE or basic residue extended forms of ME. Notably, brain levels of ME in cathepsin L-deficient mice were reduced by ≈50% compared with control mice (Fig. 7a). Relative ratios of enkephalin precursor to processed enkephalin was determined by measuring ME in tissue extracts treated with trypsin and carboxypeptidase B, which liberates ME from its precursor, compared with untreated extracts. Results (Fig. 7b) demonstrated a higher ratio of precursor/ME in brain tissue from cathepsin L KO mice compared with WT controls. These findings provide evidence for accumulation of enkephalin precursor in the cathepsin L KO mice. These findings also provide support for a functional role for cathepsin L in the production of enkephalin in brain.
Fig. 7.
Regulation of ME in cathepsin L gene KO mice. (a) ME (Met-Enk) levels in brains of KO mice. ME levels in extracts of brain tissue from cathepsin L gene KO (-/-) and WT control (+/+) mice were measured by an RIA, shown as the mean ± SEM, with 10 for each group. A significant decrease (*) in enkephalin levels in KO mice was observed (P < 0.013, two-tailed t test). (b) Ratio of precursor to enkephalin. The relative ratio of PE precursor to ME was determined by the ratio of ME measured (by an RIA) in tissue samples treated with trypsin and carboxypeptidase B compared with control untreated samples. A significant increase (*) in the ratio of precursor to ME in the KO mice was observed (P < 0.05, two-tailed t test).
In adrenals, it was of interest that ME levels in cathepsin L KO mice were increased significantly compared with WT controls (ME tissue levels were 3.25 ± 0.70 and 18.88 ± 3.52 pg of ME per mg of protein in adrenals from WT and KO mice, respectively, with P < 0.01 by t test). These findings suggest that in the absence of cathepsin L, compensatory regulatory mechanisms provided increased cellular levels of ME in adrenals. Overall, the decreased levels of ME in cortex of cathepsin L KO mice and the change in ME in adrenal together demonstrate participation of cathepsin L in regulating the production of the opioid neuropeptide ME.
Discussion
Regulated secretory vesicles of neuroendocrine chromaffin cells (known as chromaffin granules) contain ME that is generated by proteolytic processing of the PE precursor within such vesicles. The cysteine protease activity termed PTP represents the major PE-cleaving activity in secretory vesicles of chromaffin cells (7–11). In this study affinity labeling with the activity-based probe DCG-04, combined with peptide sequencing by MS, provided molecular identification of secretory vesicle cathepsin L as the cysteine protease responsible for PE-processing activity. The identified cathepsin L fulfills the criteria expected of a proneuropeptide or prohormone-processing protease (1, 2, 25). Cathepsin L, similar to native PTP activity, produced active ME by cleaving enkephalin-containing peptide substrates at paired basic and monobasic processing sites. Furthermore, cathepsin L converted full-length enkephalin precursor into intermediate products that are identical to those generated by native secretory vesicle cathepsin L (PTP) (data not shown and ref. 9). Cathepsin L colocalized with ME in regulated secretory vesicles, as observed by immunofluorescence confocal and immunoelectron microscopy. Moreover, functional cosecretion of cathepsin L with ME occurred during nicotine and KCl stimulation of chromaffin cells. Finally, cathepsin L gene KO mice possess reduced ME levels, with an increased ratio of precursor/ME in brains of cathepsin L KOs compared with WT controls.
These results demonstrate a previously uncharacterized biological function for secretory vesicle cathepsin L as a proteolytic processing enzyme for the production of active enkephalin. This newly identified function of cathepsin L in secretory vesicles contrasts with its well known function as a lysosomal protease involved in end-stage degradation of polypeptides (21). Results of this study demonstrate that cathepsin L can be routed to secretory vesicles to generate products with distinct biological functions. In chromaffin cells, the high degree of colocalization of cathepsin L immunofluorescence with ME, confirmed by dual immunoelectron microscopy, demonstrates trafficking of cathepsin L to the regulated secretory pathway. In fact, earlier studies have indicated that cathepsin L is routed to immature secretory granules of insulin-producing cells, and a significant portion of cathepsin L remains as a resident protein of the mature secretory vesicle (26). In addition, cathepsin L is present in secretory granules of pituitary GH4Cl cells and undergoes regulated secretion induced by the secretagogues forskolin and phorbol 12-myristate 13-acetate (27). Regulated secretion of cathepsin L also occurs from human lung tumor cells (28). Together, results from this study and others are consistent with a functional role for cathepsin L in secretory vesicles for producing active enkephalin and perhaps other peptides.
Evidence for a dual function of cathepsin L in secretory vesicles as well as lysosomes raises the question of how secretory vesicle cathepsin L may differ from lysosomal cathepsin L. Results from this study indicated that the molecular forms of cathepsin L in secretory vesicles differed from lysosomal cathepsin L. The secretory vesicle cathepsin L appeared as three spots of 26–28 kDa on 2D gels with DCG-04 affinity labeling; these spots differed slightly in apparent molecular masses but showed the same isoelectric points. In contrast, lysosomal cathepsin L (bovine) was composed of only one protein spot on a 2D gel, after DCG-04 labeling, of ≈27–29 kDa (S.Y. and V.H, unpublished results). In addition, the secretory vesicle cathepsin L is part of a high molecular mass complex of 180–200 kDa (10), whereas lysosomal cathepsin L is known as a single-chain form of ≈28 kDa (21). The high molecular mass components of the complex (11) have been identified by peptide sequencing with MS as β-mannosidase and dopamine β-monooxygenase. It will be of interest for future studies to examine the functional roles of the protein components of the secretory vesicle cathepsin L complex. Overall, these findings implicate different molecular forms of secretory vesicle cathepsin L compared with lysosomal cathepsin L.
It is noted that the cleavage specificity of cathepsin L in secretory vesicles, representing native PTP activity (7–10), differs somewhat from the subtilisin-like prohormone convertases known as PC1 and PC2 (2–4). Although all of these processing proteases recognize and cleave at paired basic residue sites, secretory vesicle cathepsin L (PTP) cleaves between the two residues of the dibasic site or at the NH2-terminal side of the paired basic residues. However, PC1 and PC2 preferentially cleave at the COOH-terminal side of paired basic residues within prohormones. The cleavage specificity of secretory vesicle cathepsin L (PTP) (7–9) predicts that peptide intermediate products will contain NH2-terminal basic residue extensions, which require processing by an aminopeptidase to remove these basic residues. Indeed, an aminopeptidase B-like activity is present within chromaffin granules for the removal of NH2-terminal lysine and arginine residues (29). Moreover, the presence of in vivo PE-derived intermediates in adrenal medulla with basic residue extensions at their NH2 termini (6) is consistent with the cleavage specificity of secretory vesicle cathepsin L.
Importantly, the cathepsin L gene KO mice contain reduced levels of brain ME, which represents approximately one-half of that in WT control brains. These findings substantiate a role for cathepsin L in the production of neuronal ME in brain. Adrenals showed a compensatory increase in ME in cathepsin L KO mice. Thus, in addition to the observed phenotypes of cardiomyopathy and hair loss in the cathepsin L KO mice (17, 18), these mice possess changes in enkephalin and perhaps other peptide neurotransmitters or hormones.
PE processing in brain also involves the subtilisin-like PC2 protease, because mice that contain inactive PC2 possess partial reduction of ME in brain (30, 31). The reduction of ME by approximately one-half in cathepsin L KO mice and the partial reduction of brain enkephalin in PC2-deficient mice indicate that ME production in brain involves cathepsin L as well as PC2.
In summary, results from this study provide evidence for a previously uncharacterized biological function of cathepsin L for the production of ME in the regulated secretory pathway. Secretory vesicle cathepsin L represents a newly identified proteolytic processing enzyme for the production of active enkephalin and possibly other peptide neurotransmitters or hormones.
Supplementary Material
Acknowledgments
Discussions with J. McKerrow (University of California, San Francisco) are appreciated. This research was supported by grants from the National Institutes of Health (to V.H. and B.G.) and the Sandler Program in Basic Sciences (to M.B.). K.F.M. was supported by National Institutes of Health grants to the University of California at San Francisco Mass Spectrometry Facility, directed by A. L. Burlingame.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PE, proenkephalin; PTP, prohormone thiol protease; ME, [Met]enkephalin; KO, knockout; CID, collision-induced dissociation; TOF, time-of-flight; BAM-22P, bovine adrenal medulla peptide 22; MALDI, matrix-assisted laser desorption ionization.
References
- 1.Gainer, H., Russell, J. T. & Loh, Y. P. (1985) Neuroendocrinology 40, 171-184. [DOI] [PubMed] [Google Scholar]
- 2.Hook, V. Y. H., Azaryan, A. V. & Hwang, S. R. (1994) FASEB J. 8, 1269-1278. [DOI] [PubMed] [Google Scholar]
- 3.Zhou, A., Webb, G., Zhu, X. R. & Steiner, D. F. (1999) J. Biol. Chem. 274, 20745-20748. [DOI] [PubMed] [Google Scholar]
- 4.Seidah, N. G. & Prat, A. (2002) Essays Biochem. 38, 79-94. [DOI] [PubMed] [Google Scholar]
- 5.Birch, N. P., Davie, A. D. & Christie, D. L. (1987) J. Biol. Chem. 262, 3382-3387. [PubMed] [Google Scholar]
- 6.Goumon, Y., Lugardon, K., Gadroy, P., Strub, J. M. Welters, I. D., Stefano, G. B., Aunis, D. & Metz-Boutigue, M. H. (2000) J. Biol. Chem. 275, 38355-38362. [DOI] [PubMed] [Google Scholar]
- 7.Krieger, T. K. & Hook, V. Y. H. (1991) J. Biol. Chem. 266, 8376-8383. [PubMed] [Google Scholar]
- 8.Krieger, T. K., Mende-Mueller, L. & Hook, V. Y. H. (1992) J. Neurochem. 59, 26-31. [DOI] [PubMed] [Google Scholar]
- 9.Schiller, M. R., Mende-Mueller, L., Miller, K. W. & Hook, V. Y. H. (1995) Biochemistry 34, 7988-7995. [DOI] [PubMed] [Google Scholar]
- 10.Yasothornsrikul, S., Aaron, W., Toneff, T. & Hook, V. Y. H. (1999) Biochemistry 38, 7421-7430. [DOI] [PubMed] [Google Scholar]
- 11.Tezapsidis, N., Noctor, S., Kanna, R., Krieger, T. J., Mende-Mueller, L. & Hook, V. Y. H. (1995) J. Biol. Chem. 270, 13285-13290. [DOI] [PubMed] [Google Scholar]
- 12.Law, P. Y., Wong, Y. H. & Loh, H. H. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 389-430. [DOI] [PubMed] [Google Scholar]
- 13.Pasternak, G. W. (1993) Clin. Neuropharmacol. 16, 1-18. [DOI] [PubMed] [Google Scholar]
- 14.Greenbaum, D., Medzihradszky, K. F., Burlingame, A. & Bogyo, M. (2000) Chem. Biol. 7, 569-581. [DOI] [PubMed] [Google Scholar]
- 15.Greenbaum, D., Arnold, W., Lu, F., Hayrapetian, L., Baruch, A., Krumrine, J., Toba, S., Chehade, K., Bromme, D., Kuntz, I. D. & Bogyo, M. (2002) Chem. Biol. 9, 1085-1094. [DOI] [PubMed] [Google Scholar]
- 16.Reinheckel, T., Deussing, J. Roth, W. & Peters, C. (2001) Biol. Chem. 382, 735-741. [DOI] [PubMed] [Google Scholar]
- 17.Tobin, D. J., Foitzik, K., Reinheckel, T., Mecklenburg, L., Botchkarev, V. A., Peters, C. & Paus, R. (2002) Am. J. Pathol. 160, 1807-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stypmann, J., Glaser, K., Roth, W., Tobin, D. J., Petermann, I., Matthias, R., Monnig, G., Haverkamp, W., Breithardt, G., Schmahl, W., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 6234-6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hook, V., Noctor, S., Sei, C., Toneff, T., Yasothornsrikul, S., Byrne, M., Tezapsidis, N., Johnston, J. & Kang, Y. H. (1999) Endocrinology 140, 3744-3754. [DOI] [PubMed] [Google Scholar]
- 20.Hwang, S. R., Steineckert, B., Toneff, T., Bundey, R., Logvinova, A. V., Goldsmith, P. & Hook, V. Y. H. (2002) Biochemistry 41, 10397-10405. [DOI] [PubMed] [Google Scholar]
- 21.Barrett, A. J., Rawlings, N. D. & Woessner, J. F. (1998) in Handbook of Proteolytic Enzymes (Academic, San Diego), pp. 609-617.
- 22.Greenbaum, D., Baruch, A., Hayrapetian, L., Darula, Z., Burlingame, A., Medzihradszky K. F. & Bogyo, M. (2002) Mol. Cell. Proteomics 1, 60-68. [DOI] [PubMed] [Google Scholar]
- 23.Gour-Salin, B. J., Lachance, P., Ploufe, C., Storer, A. C. & Menard, R. (1993) J. Med. Chem. 36, 720-725. [DOI] [PubMed] [Google Scholar]
- 24.Bogyo, M., Verhelst, S., Bellingard-Dubouchaud, V., Toba, S. & Greenbaum, D. (2000) Chem. Biol. 1, 27-38. [DOI] [PubMed] [Google Scholar]
- 25.Docherty, K. & Steiner, D. F. (1982) Annu. Rev. Physiol. 44, 625-638. [DOI] [PubMed] [Google Scholar]
- 26.Kuliat, R., Klumperman, J., Ludwig, T. & Arvan, P. (1997) J. Cell Biol. 137, 595-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waguri, S., Sato, N., Watanbe, T., Ishidoh, K., Kominami, E., Sato, K. & Uchiyama, Y. (1995) Eur. J. Cell Biol. 67, 308-318. [PubMed] [Google Scholar]
- 28.Ulbricht, B., Henny, H., Horstmann, H., Spring, H., Faigle, W. & Spiess, E. (1997) Eur. J. Cell Biol. 74, 294-301. [PubMed] [Google Scholar]
- 29.Yasothornsrikul, S., Toneff, T. & Hook, V. Y. H. (1998) J. Neurochem. 70, 153-163. [DOI] [PubMed] [Google Scholar]
- 30.Johanning, K., Juliano, M. A., Juliano, L., Lazure, C., Lamango, N. S., Steiner, D. F. & Lindberg, I. (1998) J. Biol. Chem. 273, 22672-22680. [DOI] [PubMed] [Google Scholar]
- 31.Miller, R., Toneff, T., Vishnuvardhan, D., Beinfeld, M. & Hook, V. Y. H. (2003) Neuropeptides 37, 140-148. [DOI] [PubMed] [Google Scholar]
- 32.Biemann, K. (1990) Enzymology 193, 886-887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.