Abstract
Cytoplasmic polyadenylation element-binding (CPEB) proteins control polyadenylation-induced translation in early development. Studies in oocytes led to the delineation of Xenopus CPEB, the first member of the family to be identified, and its mouse homologue mCPEB-1. Recently, a second mouse family member, mCPEB-2, has been described in germ cells. Increasing evidence also implicates CPEB proteins as being important in the hippocampus, where these proteins are thought to regulate local protein synthesis and synaptic plasticity. We therefore carried out a systematic screen for CPEB genes in the mouse brain and report two previously undescribed gene family members: mCPEB-3 and -4. We next examined the expression of all four genes in the hippocampus and found that mCPEB-1, -2, and -4 transcripts are expressed in the principal cell layer in the CA3 and CA1 region and in the dentate gyrus of the hippocampus. mCPEB-3 was barely expressed in naïve animals but together with mCPEB-4 was strongly up-regulated after injection of kainate to initiate seizure activity. Whereas mCPEB-1 is regulated by the Aurora kinase, mCPEB-2, -3, and -4 do not contain Aurora kinase phosphorylation sites. However, alternative splice isoforms of mCPEB-2, -3, and -4 encode the so-called B region with phosphorylation sites for cAMP-dependent protein kinase, calcium/calmodulin-dependent protein kinase II, and S6 kinase. Only isoforms that encode the B region were expressed in the principal cell layer. Coexpression of mCPEB-1 and the B region-containing splice isoforms suggests that a variety of different signaling pathways can recruit CPEB activity in hippocampal neurons.
Local protein synthesis has long been known to be important for temporal and spatial control of embryonic development (1) and has recently also been shown to be important in synaptic plasticity (2–4). Several transcripts crucial to these changes possess 3′ UTRs with polyadenylated tails that can be subject to stimulus-dependent elongation by cytoplasmic polyadenylation (1, 4–8). Transcripts so regulated contain a conserved nucleotide sequence in the 3′ UTR: the cytoplasmic polyadenylation element (CPE) with the consensus UUUUUAU (1). These sequences are recognized by the CPE-binding protein (CPEB), which aids in the assembly of a protein complex that allows polyadenylation (1). CPEB is not only an activator, however; it is also capable of acting as a repressor of translation. Indeed, under basal conditions, CPEB keeps bound mRNA in a dormant state (1). In the case of CPEB-1, this repression is relieved by phosphorylation (9) or degradation (10) of CPEB. As a result, local signals acting on CPEB contribute to the spatial specificity of CPEB-mediated local protein synthesis (11).
Much of the classical initial work on CPEB-1 was carried out in Xenopus oocytes. However, recently CPEB-1 has also been found to be present in the hippocampus (7), where protein synthesis is essential for the late phase of long-term potentiation (L-LTP) (12, 13). In the rodent CNS, activation of Aurora kinase by the N-methyl-d-aspartate receptor leads to the phosphorylation of CPEB-1 and the subsequently enhanced cytoplasmic polyadenylation and translation of the transcript encoding the α subunit of the calcium/calmodulin-dependent protein kinase (CaMKIIα) (14), a key element in hippocampal LTP (15, 16). The 3′ UTR of CaMKIIα transcripts regulates dendritic targeting of the mRNA (17) and contains CPE elements (7). Indeed, mice lacking the CaMKIIα 3′ UTR show strongly decreased dendritic mRNA localization, a decreased protein content in postsynaptic density fractions, and a deficit in L-LTP (18). However, electrophysiological investigation of mCPEB-1-deficient mice (19) revealed only modest changes in L-LTP (J. M. Alarcon, R. Hodgman, M.T., E.R.K., and J. D. Richter, unpublished data). Because lack of mCPEB-1 may be compensated for by other proteins that mediate local protein synthesis, we were prompted to search for additional CPEB family members expressed in mouse brain. Recently, the murine isoform mCPEB-2 was found in testis (20). In addition, two human brain cDNAs, called KIAA0940 and KIAA1673, have been identified as potentially encoding CPEB proteins (1) with high homology to mCPEB-2 (20).
We have now characterized the expression of mCPEB-2 in the brain. In addition, we have isolated two previously undescribed members (mCPEB-3 and -4) in the mouse and compared their expression pattern in the brain to that of mCPEB-2 and -1. mCPEB-3 and -4 were strongly induced in vivo by injection of kainate, a glutamate receptor agonist that leads to strong neuronal activation and seizures (21). Unlike mCPEB-1, the other CPEB isoproteins lack Aurora kinase phosphorylation sites. In the so-called B region, whose presence depends on alternative splicing, mCPEB-2, -3, and -4 possess putative phosphorylation sites for cyclic AMP-dependent protein kinase (PKA), CaMKII, and p70S6 kinase, a growth-factor-stimulated serine threonine kinase that acts on components of the translational apparatus (22). In the hippocampus, splice isoforms that possess the B region encoding these phosphorylation sites were enriched in the principal cell layers and might possibly compensate for mCPEB-1 deficiency.
Materials and Methods
Amplification of cDNAs and Generation of Full-Length ORFs. Mouse brain cDNA (Marathon-Ready and QuickClone, CLONTECH; First Choice, Ambion, Austin, TX) was amplified by using the Advantage2 PCR kit (CLONTECH). PCR applications and primer sequences can be found in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org. For sequence comparisons, the clustalw method and megalign program (DNASTAR, Madison, WI) were used.
Detection of mCPEB Isoforms by PCR. Mouse brain cDNA and plasmid preparations of individual splice isoforms were subjected to PCR by using the AccuPrime kit (Invitrogen). Determination of mCPEB-3 splice forms according to fragment size was performed (60°C, 1 min) by using primers CCATGGCTGTCATCCAAGAAGGCGTC and GGATATGATAAGGACTGACCATGAGCCTCTGAAAG. Primers CAGACCACTA TGAAGAGGTTGATCCCCACG and CCCAGGACGTTTGACATGCACTCACTG, spanning the variable region, were used (60°C, 1 min) to distinguish mCPEB-4 splice isoforms by fragment size.
Northern Blot Hybridization. Mouse tissue array Northern blots were processed according to the manufacturer's instructions (First Choice and Strip-EZ DNA kit, catalog no. 1470B4, Ambion). Probes for the N-terminal coding regions of the respective mCPEB isoforms were between 320 and 390 nt large. For mCPEB-1, a 388-bp EcoRI/BssSI fragment was used, whereas mCPEB-2 transcripts were detected by using a 322-bp BglII/DraI probe. For mCPEB-3, we probed with a 336-bp fragment amplified from brain cDNA by using primers CGGGTTGACGTGGTGCGGGAAGTTTTG and GAAGCCCCGTCCACGCCCCTCTCCTCAG and a 360-bp BglII/Eco0109I fragment specifically hybridized to mCPEB-4 transcripts. Human KIAA0940 transcripts were detected by using a human multiple tissue Northern blot (CLONTECH, no. 7780–1). A 302-bp probe for the proximal 3′ UTR was amplified by using primers TGATTCTTTTGTTTGTTGTTGTGG and CCTCGGCAAAACAAAAATCAAACA.
In Situ Hybridization with End-Labeled Oligonucleotides. In situ hybridization on brain cryosections was performed as described (23). Oligonucleotide sequences are in Supporting Text. For induction of neuronal gene expression, mice were injected i.p. with kainic acid (Sigma) dissolved in PBS (20 mg/kg body weight) and housed in individual cages. Seizure activity was monitored. Mice were killed one (n = 4), 2 (n = 4), 4 (n = 6), and 8 h (n = 3) after injection. Noninjected mice (n = 6) served as control. Brains were embedded in Tissue Tek (Sakura Finetek, Torrance, CA), fresh frozen on dry ice, and stored at –70°C.
Results
Only one isoform of CPEB has been described in mouse brain previously. In the search for other isoforms, we performed PCR analysis of mouse brain cDNA with primers obtained from mouse sequences highly similar to human cDNAs encoding CPEB-like proteins. We found three other CPEB isoforms also expressed in brain, each of which had several splice variants. In addition to mCPEB-1 and -2, whose sequences have been previously delineated, we found the new isoforms mCPEB-3 and -4.
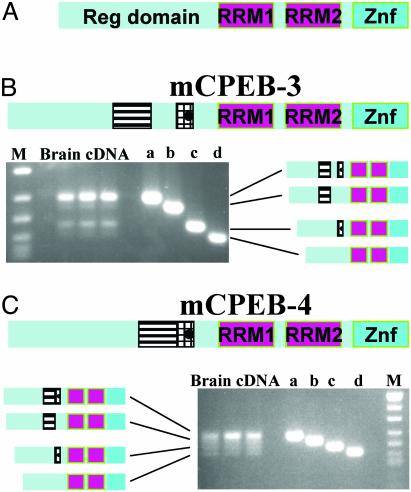
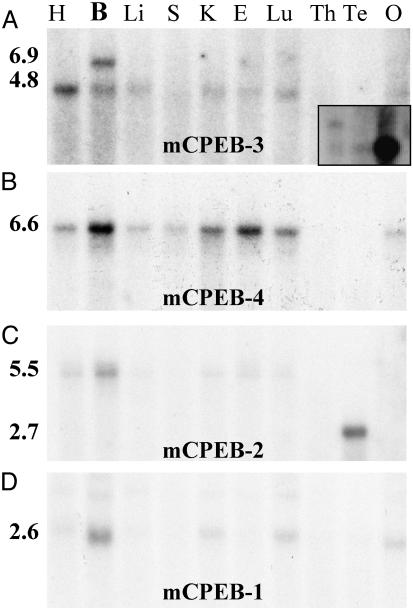
Isolation and Transcript Distribution of mCPEB-3a–d. A 3,075-bp fragment carrying the full-length ORF of the murine KIAA0940 homologue (called mCPEB-3) and part of the 3′ UTR (GenBank accession no. AY313774) was obtained from mouse brain cDNA. The isolated cDNA encoded a polypeptide that showed 97.5% sequence identity to the hypothetical protein (NP_055727.1) encoded by the human KIAA0940 cDNA from brain and matched with putative exons on mouse chromosome 19 (Ensemble database; www.ensembl.org/Mus_musculus). Sequencing revealed four different splice isoforms (Figs. 1 and 2B). The longest isoform of mCPEB-3 (called mCPEB-3a) encoded a polypeptide of 716-aa length, whereas the mCPEB-3b isoform had a 24-bp deletion resulting in an 8-aa deletion (B deletion). The mCPEB-3c isoform was characterized by a large deletion of 69 bp (23 aa, C deletion), and the mCPEB-3d isoform lacked both sequences, i.e., 93 bp (31 aa). Semiquantitative PCR with primers flanking the alternatively spliced regions revealed that the mCPEB-3a isoform was most abundant in brain (Fig. 2B). We performed 3′ RACE of brain cDNA, which revealed a short 3′ UTR of 927 bp and a long brain-specific 3′ UTR of ≈3.5 kb, both of which contained CPEs near a polyadenylation signal. Partial sequencing of the large RACE product showed high sequence identity to the terminal sequences of human KIAA0940 cDNA (NM_014912, not shown). Northern blot hybridization for mCPEB-3 mRNA revealed strong expression in heart and brain (Fig. 3A). Two different transcript sizes for mCPEB-3 were found in the brain: a brain-specific long transcript of 6.9 kb and a ubiquitous transcript of 4.8 kb. Weaker expression of the 4.8-kb transcript was found in liver, kidney, embryo, lung, and ovary. Both mRNA sizes are consistent with the presence of short and long 3′ UTRs found by RACE of brain cDNA. When we determined the transcript distribution for the human KIAA0940, we also found a brain-specific long transcript in addition to a smaller transcript in brain, heart, and skeletal muscle (see Fig. 3A Inset).
Fig. 1.
Deduced amino acid sequences for mCPEB-3 and -4. A glutamine-rich region is shown in italics. RNA recognition motifs (RRMs) and conserved histidine and cysteine residues from the Zn-finger domain characteristic for CPEB proteins (1) are underlined. Amino acids encoded by B exons are shown in boldface; amino acids encoded by the C exons are shown in boldfaced italics. The corresponding nucleotide sequences are available at GenBank.
Fig. 2.
mCPEB-3 and -4 splice isoforms in mouse brain. (A) CPEB proteins contain an N-terminal regulatory domain and a C-terminal RNA-binding domain that consists of two RNA recognition motifs and a Zn-finger domain (Znf). (B) Schematic view of mCPEB-3 and relative quantitation of different splice isoforms. (Upper) mCPEB-3 contains a variable region with two amino acid stretches that are lacking in some splice variants. The B stretch (hatched box) contains putative phosphorylation sites (filled circle) and is separated from the C stretch (striated box). (Lower Left) Analysis of brain cDNA subjected to PCR with primers flanking the variable region yields four amplicons differing in size and intensity. Corresponding plasmids containing the different splice isoforms (a–d) are run in parallel. M, 100-base-pair ladder. (Lower Right) Schematic view of the corresponding variant polypeptides. The mCPEB-3a splice isoform is most abundant in brain. (C) Presence of mCPEB-4 isoforms in brain. For explanations, see B.(Upper) Variable region in the N-terminal half. Unlike mCPEB-3, the B stretch is not separated from the C stretch. (Lower Right) PCR with primers flanking the variable region yields four amplicons differing in size and intensity. Corresponding plasmids containing the four mCPEB-4 splice isoforms served as size standards. (Lower Left) Schematic view of the corresponding variant polypeptides. The mCPEB-4a isoform is slightly more abundant in brain.
Fig. 3.
Tissue distribution of mouse CPEB transcripts determined by Northern blotting. Tissue array Northern blots were hybridized to probes specific for mCPEB-3 (A), -4 (B), -2 (C), and -1 (D). Transcript sizes are indicated in kilobases. H, heart; B, brain; Li, liver; S, spleen; K, kidney; E, embryo 14.5 days postcoitum; Lu, lung; Th, thymus; Te, testis; O, ovary. (A Inset) Northern blot for human KIAA0940 shows strong expression in skeletal muscle (Right) and weaker expression in heart (Center), and brain (Left), which shows a brain-specific long transcript as observed in mouse.
Isolation and Transcript Distribution of mCPEB-4a–d. We amplified the full-length coding region (GenBank accession no. AY313775) of the murine KIAA1673 protein homologue mCPEB-4 from mouse brain cDNA. The isolated cDNA encoded a polypeptide with 98.2% sequence identity to the protein (XP_047672.4) deduced from the human KIAA1673 cDNA from brain. Sequencing of individual plasmid preparations revealed four different splice isoforms (Figs. 1 and 2C). The longest isoform of mCPEB-4 (called mCPEB-4a) encoded a polypeptide of 729-aa length, whereas the mCPEB-4b isoform had a 24-bp deletion (8 aa, B deletion). The mCPEB-4c isoform was characterized by a large deletion of 51 bp (17 aa, C deletion), and the mCPEB-4d isoform lacked both sequences, i.e., 75 bp (25 aa). The cDNA sequences corresponded to putative exons on mouse chromosome 11 (Ensembl database). Semiquantitative PCR of three different mouse brain cDNAs with primers flanking the alternatively spliced region yielded four amplicons. The mCPEB-4a isoform seemed most abundant (Fig. 2C). Northern blot hybridization revealed the strongest expression of a 6.6-kb mCPEB-4 transcript in brain and in 14.5-day postcoitum embryos; strong expression in kidney, lung, and heart; and weaker expression in liver, spleen, and ovary (Fig. 3B).
Isolation and Expression Analysis of mCPEB-2. mCPEB-2 has recently been isolated from testis. We amplified a fragment of the mCPEB-2 ORF from mouse brain cDNA. The isolated product showed 100% sequence identity to the recently described mCPEB-2 mRNA from testis (20) and the cDNA XM_132072. Both published sequences differed by a 24-bp deletion in testis cDNA. The deletion was homologous to the B deletion of mCPEB-3 and -4 (Fig. 4B). Using 5′ RACE, we extended the sequence information from brain cDNA by 339 bp and always obtained sequences encoding the B region. Another difference compared with the mCPEB-2 mRNA from testis was an extension of several kilobases in the 3′ UTR of XM_132072. Using 3′ RACE of brain cDNA, we obtained a large 4.5-kb product. The terminal sequences were identical to the 3′ end of XM_132072. The calculated length of the 3′ UTR was ≈3.8 kb, longer than the 3′ UTR described in the mCPEB-2 mRNA from testis (20). Northern blot hybridization for mCPEB-2 mRNA showed a 2.7-kb fragment in testis (Fig. 3C) that has been described (20). We additionally observed a long 5.5-kb transcript with strongest expression in brain and weaker expression in heart, kidney, embryo, and lung (Fig. 3C). Thus, mCPEB-2 has a short transcript that is very abundantly expressed in testis and a long transcript that is expressed at a lower level in other tissues.
Fig. 4.
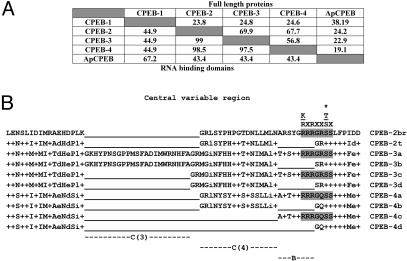
Comparison of CPEB family members. (A) Homology percentages of the full-length proteins and of the RNA-binding domains were calculated by using clustalw, respectively, for the mouse and Aplysia CPEBs. (B) Sequence comparison of variable regions in mCPEB-2, -3, and -4 proteins. The B region is conditionally present in all mCPEBs. Consensus phosphorylation sites for PKA, CaMKII, and p70S6 kinase are shown above the sequences, and the phosphorylated residue is marked by an asterisk. The corresponding actual recognition sites are shaded. The a and c isoforms of mCPEB-3 and -4 and the mCPEB-2 isoform isolated from brain (mCPEB-2br) possess this site. Note that the phosphorylated serine residue does not reside in the variable region. However, the kinase recognition sites are disrupted by the B deletion. The b and d isoforms of mCPEB-3 and -4 and the testis-specific mCPEB-2 isoform (mCPEB-2t) lack the B region (underlined). Only mCPEB-4 isoforms conditionally lack the C (4) region. Only mCPEB-3 isoforms conditionally lack the C (3) region. +, conserved residues. Similar residues are written in lower case; gaps are underlined.
Finally, we determined the tissue distribution of mCPEB-1 mRNA by Northern blotting (Fig. 3D). We found a transcript of ≈2.6 kb most abundant in brain, which also showed strong expression in kidney, lung, and ovary and weaker expression in heart.
Comparison of the Mouse CPEB Family Members. The mouse mCPEB-3 and -4 isoforms, homologues of human KIAA0940 and KIAA1673, are most similar to mCPEB-2 and show less homology to mCPEB-1 and Aplysia CPEB (Fig. 4A). The similarity among the three mouse subfamily members is strongest in the RNA-binding domain (Fig. 4A). We identified a central region that was characterized by modest sequence homology and interspersed variations, i.e., insertions and deletions (Fig. 4B). All full-length mCPEB proteins contained an 8-aa stretch called the B region with the consensus sequence T/ART/SYGRRR. The region was lacking in mCPEB-2 from testis and in the b and d isoforms of mCPEB-3 and -4 (Fig. 4B). We analyzed mCPEB polypeptides for the presence of Aurora kinase phosphorylation sites as described (10) and for additional phosphorylation sites by using the web tools netphos 2.0 (www.cbs.dtu.dk/services/NetPhos) and phosphobase (www.cbs.dtu.dk/databases/PhosphoBase). In contrast to mCPEB-1, which has Aurora kinase phosphorylation sites, the deduced mCPEB-2, -3, and -4 polypeptides did not contain Aurora kinase phosphorylation sites. However, for all those mCPEBs, we found a site within the B variable region (Fig. 4B) that provides consensus recognition sites for phosphorylation by PKA and CaMKII (R-X-X-S/T-X; refs. 24 and 25) and p70S6 kinase (K/R-X-RX-X-S/T-X; ref. 26). These sites allow phosphorylation of a serine residue adjacent to the B region exclusively in a and c isoforms of mCPEB-3, -4, and -2 from brain. However, those recognition sites are not universal and are absent in b and d isoforms of mCPEB-3, -4, and -2 from testis (Fig. 4B).
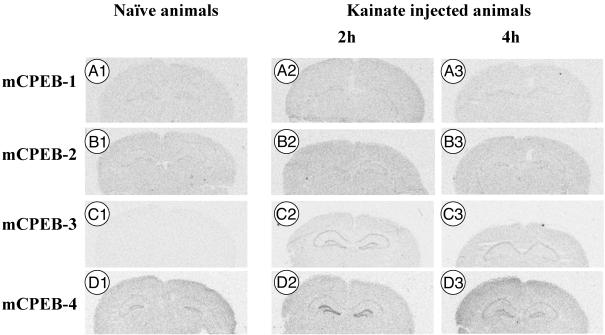
Cell-Type Specificity in Brain. We determined mCPEB-2, -3, and -4 expression by in situ hybridization of mouse brain and compared their expression pattern with mCPEB-1 (Fig. 5A1). Whereas mCPEB-4 (Fig. 5D1) showed a higher basal expression level compared with mCPEB-1 in the principal cells of the hippocampal formation, mCPEB-3 was barely detectable (Fig. 5C1). mCPEB-2 showed expression in principal cells of the hippocampus (Fig. 5B1) with intensity and distribution similar to mCPEB-1.
Fig. 5.
mCPEB transcript distribution in mouse hippocampus under basic conditions and after kainate induction. Coronal cryosections were hybridized with oligonucleotides specific for mCPEB isoforms. (A1–A3) mCPEB-1. (B1–B3) mCPEB-2. (C1–C3) mCPEB-3. (D1–D3) mCPEB-4. Expression of mCPEB isoforms is shown under basic conditions (A1, B1, C1, and D1), 2 h after kainate induction (A2, B2, C2, and D2), and 4 h after kainate induction (A3, B3, C3, and D3). Exposition was 3 days for mCPEB-3 and -4 probes and 4 wk for mCPEB-1 and -2 probes. Except for mCPEB-2, two probes were used for each transcript, yielding qualitatively identical results.
With the exception of mCPEB-4, the expression levels of the mCPEBs, as determined by in situ hybridization, were low in the normal mouse brain. We therefore tested whether mCPEBs were induced by strong neuronal stimulation, injected kainate i.p., and analyzed brains at different time points after induction compared with noninjected control animals. As a positive control, we used Arc, an mRNA known to be induced by electrical induction of seizures (27). Arc mRNA was hardly detectable in the basal state but strongly up-regulated 1, 2, 4, and 8 h after kainate stimulation (not shown). mCPEB-3 was barely detectable in the basal state (Fig. 5C1) and 1 h postinjection (Table 1) but was strongly expressed in the principal cells of the cornu ammonis and dentate gyrus 2 h after injection (Fig. 5C2). The expression of mCPEB-3 in dentate gyrus was decreased to basal levels 4 h postinjection in four of six animals but remained stable in CA3 and CA1 (Fig. 5C3). Eight hours later, expression decreased but remained elevated in CA1/CA3, albeit at lower levels than at 4 and 2 h (Table 1). In parallel, mCPEB-4 was up-regulated 1 h (Table 1) and 2 h after injection in granule cells of the dentate gyrus (Fig. 5D2) and was decreased to levels slightly above basal levels in dentate gyrus 4 h after stimulation (Fig. 5D3). In CA3 and CA1, up-regulation was apparent 4 h after stimulation (Fig. 5D3). After 8 h, expression reached basal levels (Table 1). In contrast, mCPEB-1 levels did not change at all during stimulation (Fig. 5A1–3) and were low compared with the basal level of mCPEB-4 (Fig. 5D1) and the induced mCPEB-3 levels (Fig. 5 C2 and C3). Similarly, mCPEB-2 was not influenced by kainic acid (Fig. 5B1–3).
Table 1. Relative mCPEB transcript abundance in the principal cell layers of the hippocampus at different time points after kainate injection.
| Hours | CPEB-1 | CPEB-2 | CPEB-3 | CPEB-4 |
|---|---|---|---|---|
| CA3/1 | ||||
| 0 | + | + | - | ++ |
| 1 | + | + | - | ++ |
| 2 | + | + | +++ | ++ |
| 4 | + | + | +++ | +++ |
| 8 | + | + | + | ++ |
| DG | ||||
| 0 | + | + | - | ++ |
| 1 | + | + | - | ++++ |
| 2 | + | + | +++ | ++++ |
| 4 | + | + | - | ++(+) |
| 8 | + | + | - | ++ |
Staining is graded as barely detectable (-), low (+), moderate (++), strong (+++), and very strong (++++). DG, dentate gyrus
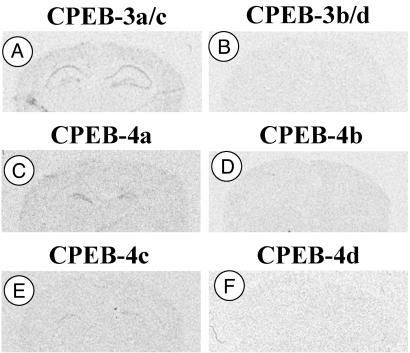
To determine the isoform specificity of mCPEB expression in the principal cell layers of the hippocampus, we used oligonucleotides complementary to DNA sequences flanking the individual splice sites of mCPEB-3 and -4. For mCPEB-3, it was not possible to derive oligonucleotides exclusively directed to individual isoforms, because the C and B exons are separated by an exon that is contained in all mCPEB-3 isoforms. Starting 2 h after induction, we found expression in principal cells by using an oligonucleotide directed to both the mCPEB-3a and -3c isoforms that contain the B region (Fig. 6A) but not with another oligonucleotide directed to both the mCPEB-3b and -3d isoforms that lack the B region (Fig. 6B). For mCPEB-4, we obtained labeling of the principal cell layers of the hippocampus 1 h after induction by using an oligonucleotide specific for mCPEB-4a (Fig. 6C) and weak labeling with an oligonucleotide specific for mCPEB-4d (Fig. 6E). The two isoforms that lack the B exon encoding the putative phosphorylation sites (mCPEB-4b and -4d) were not detected in the principal cell layers either in naïve animals or after induction (Figs. 6 D and F).
Fig. 6.
Isoform distribution of mCPEB-3 and -4 in principal cell layers of the hippocampus. (A) mCPEB-3a/c-specific probe, 2 h after kainate induction. (B) mCPEB-3b/d-specific probe (2 h). (C) mCPEB-4a-specific probe (1 h). (D) mCPEB-4b-specific probe (1 h). (E) mCPEB-4c-specific probe (1 h). (F) mCPEB-4d-specific probe (1 h).
Discussion
The modest changes in the synaptic physiology in mCPEB-1-deficient mice (J. M. Alarcon, R. Hodgman, M.T., E.R.K., and J. D. Richter, unpublished data) and the recent observation in Aplysia that there might be neuron-specific isoforms of CPEB (5) prompted us to carry out a systematic search for new CPEB family members in mice that might be present in brain. We now have identified two putative mouse CPEB isoforms, which we call mCPEB-3 and -4. These two differ from mCPEB-1 both in their patterns of expression and in their regulatory features. This raises the possibility that the different family members of CPEB might regulate different aspects of the temporal and spatial order of local protein synthesis.
Mouse CPEB Family Members Have Distinct Neuronal Expression Patterns. We found that all four CPEB genes are expressed in granular cells of dentate gyrus and the pyramidal cells of CA3 and CA1. However, unlike mCPEB-1 and -2, the isoforms mCPEB-3 and -4 were up-regulated by strong neuronal activation. Interestingly, the up-regulation of mCPEB-3 and -4 is transient in the dentate gyrus but more long lasting in the CA3 and CA1 region. Their spatiotemporal kinetics are intriguing because they follow the principal path of the information flow in the hippocampus (28). The complexity of expression pattern is further accentuated by the differential distribution of various splice isoforms.
The mouse mCPEB-3 protein is particularly interesting, because it seems a likely candidate for being the functional homologue of the Aplysia neuronal CPEB. mCPEB-3 shares three common features with Aplysia CPEB. First, in the N-terminal domain, both possess a glutamine-rich region (ref. 5; Fig. 1). Second, neither of them contains Aurora kinase phosphorylation sites. Finally, the expression of both Aplysia CPEB and mouse mCPEB-3 is up-regulated after neuronal stimulation: for Aplysia CPEB, regulation occurs posttranscriptionally (ref. 5; K.S., M. Giustetto, A. Etkin, R. Hsu, H. Zhu, and E.R.K., unpublished data), whereas mCPEB-3 is up-regulated at the transcriptional level (Fig. 5). A conceivable late activity of mCPEB-3 protein supporting expression of L-LTP or its maintenance would be consistent with the observed late induction of mCPEB-3.
Mouse CPEB Family Members Might Be Regulated by Distinct Signaling Pathways. What controls the activity of different mouse CPEB family members? In the case of mCPEB-1, stimulation of hippocampal cells by glutamate leads to the N-methyl-d-aspartate-receptor-dependent activation of Aurora kinase, which in turn phosphorylates mCPEB-1 (14). In contrast to mCPEB-1, the other mouse CPEB family members do not contain Aurora kinase phosphorylation sites. Instead, all of them have splice variants that are selectively expressed in the principal cell layers and contain the putative recognition sites for PKA, CaMKII, and S6 kinase. Interestingly, S6 kinase is a downstream effector of FRAP/mTOR kinase pathway (22), a pathway that has already been implicated in L-LTP (29). An inhibitor of FRAP/mTOR, rapamycin, blocks the late phase of LTF in Aplysia and L-LTP in the mouse (2, 29). CPEB activity might also be regulated by PKA (5) or CaMKII phosphorylation. In addition, mCPEBs might control their expression by an autoregulatory feedback loop: RACE experiments for mCPEB-3 yielded two different transcripts, both of which contain putative CPEs near polyadenylation signals. The Aplysia homologue of mCPEB-3 also contains CPEs in its 3′ UTR (K.S., M. Giustetto, A. Etkin, R. Hsu, H. Zhu, and E.R.K., unpublished data). This raises the possibility that activation of mCPEB-1 after N-methyl-d-aspartate receptor activation (7, 14) can in turn result in the activation of mCPEB-3. The autoregulatory loop of CPEB activation has already been demonstrated in Drosophila (30).
Different mCPEB Molecules Might Be Involved in Different Phases or Forms of Synaptic Plasticity. Why are there so many mCPEB molecules, and what specific role do they play in long-term synaptic plasticity? That mCPEB1, -2, and -4 are present in the basal state and mCPEB-3 and -4 are induced sequentially may reflect a differential temporal requirement of mCPEBs. Alternatively, different mCPEBs might be involved as well in distinct forms of synaptic plasticity or in nonplastic functions. A clue to such possibility came from the analysis of LTP in mCPEB-1-deficient mice: LTP induced by the neurotrophin brain-derived neurotrophic factor (BDNF) was completely unaffected by the absence of mCPEB-1 (J. M. Alarcon, R. Hodgman, M.T., E.R.K., and J. Richter, unpublished data), even though BDNF-mediated LTP requires local protein synthesis (29). What might be controlling the local synaptic protein synthesis in BDNF-dependent LTP? Because rapamycin completely abrogates BDNF-mediated LTP (29) and mCPEB-2, -3, and -4 contain a putative S6 kinase site, one can imagine part of this rapamycin-sensitive translational activation can be mediated by mCPEB-3 and -4. Indeed the induction of the Aplysia homologue of mCPEB-3 is blocked by rapamycin (K.S., M. Giustetto, A. Etkin, R. Hsu, H. Zhu, and E.R.K., unpublished data). Finally, different mCPEB family members might have distinct substrate specificities.
Note Added in Proof. For the human CPEB gene family, a nomenclature has been proposed (10) that differs from the nomenclature for the mouse CPEB gene family (20).
Supplementary Material
Acknowledgments
We thank James Schwartz for critical comments. This work was supported by the Howard Hughes Medical Institute.
Abbreviations: CPE, cytoplasmic polyadenylation element; CPEB, CPE-binding protein; mCPEB, mouse homologue of CPEB; PKA, cyclic AMP-dependent protein kinase; CaMK, calcium/calmodulin-dependent protein kinase; LTP, long-term potentiation; L-LTP, late-phase LTP.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY313774 for mCBEP-3 and AY313775 for mCPEB-4).
References
- 1.Mendez, R. & Richter, J. D. (2001) Nat. Rev. Mol. Cell Biol. 2, 521-529. [DOI] [PubMed] [Google Scholar]
- 2.Casadio, A., Martin, K. C., Giustetto, M., Zhu, H., Chen, M., Bartsch, D., Bailey, C. H. & Kandel, E. R. (1999) Cell 99, 221-237. [DOI] [PubMed] [Google Scholar]
- 3.Martin, K. C., Barad, M. & Kandel, E. R. (2000) Curr. Opin. Neurobiol. 10, 587-592. [DOI] [PubMed] [Google Scholar]
- 4.Steward, O. & Schuman, E. M. (2001) Annu. Rev. Neurosci. 24, 299-325. [DOI] [PubMed] [Google Scholar]
- 5.Liu, J. & Schwartz, J. H. (2003) Brain Res. 959, 68-76. [DOI] [PubMed] [Google Scholar]
- 6.Wells, D. G., Dong, X., Quinlan, E. M., Huang, Y. S., Bear, M. F., Richter, J. D. & Fallon, J. R. (2001) J. Neurosci. 21, 9541-9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu, L., Wells, D., Tay, J., Mendis, D., Abbott, M. A., Barnitt, A., Quinlan, E., Heynen, A., Fallon, J. R. & Richter, J. D. (1998) Neuron 21, 1129-1139. [DOI] [PubMed] [Google Scholar]
- 8.Minshall, N., Walker, J., Dale, M. & Standart, N. (1999) RNA 5, 27-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendez, R., Hake, L. E., Andresson, T., Littlepage, L. E., Ruderman, J. V. & Richter, J. D. (2000) Nature 404, 302-307. [DOI] [PubMed] [Google Scholar]
- 10.Mendez, R., Barnard, D. & Richter, J. D. (2002) EMBO J. 21, 1833-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richter, J. D. & Lorenz, L. J. (2002) Curr. Opin. Neurobiol. 12, 300-304. [DOI] [PubMed] [Google Scholar]
- 12.Frey, U., Krug, M., Reymann, K. G. & Matthies, H. (1988) Brain Res. 452, 57-65. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen, P. V., Abel, T. & Kandel, E. R. (1994) Science 265, 1104-1107. [DOI] [PubMed] [Google Scholar]
- 14.Huang, Y. S., Jung, M. Y., Sarkissian, M. & Richter, J. D. (2002) EMBO J. 21, 2139-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamoto, E. & Fukunaga, K. (1996) Neurosci. Res. 24, 117-122. [DOI] [PubMed] [Google Scholar]
- 16.Silva, A. J., Stevens, C. F., Tonegawa, S. & Wang, Y. (1992) Science 257, 201-206. [DOI] [PubMed] [Google Scholar]
- 17.Mayford, M., Baranes, D., Podsypanina, K. & Kandel, E. R. (1996) Proc. Natl. Acad. Sci. USA 93, 13250-13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, S., Yasuda, M., Coats, J. K., Jones, Y., Martone, M. E. & Mayford, M. (2002) Neuron 36, 507-519. [DOI] [PubMed] [Google Scholar]
- 19.Tay, J. & Richter, J. D. (2001) Dev. Cell 1, 201-213. [DOI] [PubMed] [Google Scholar]
- 20.Kurihara, Y., Tokuriki, M., Myojin, R., Hori, T., Kuroiwa, A., Matsuda, Y., Sakurai, T., Kimura, M., Hecht, N. B. & Uesugi, S. (2003) Biol. Reprod. 69, 261-268. [DOI] [PubMed] [Google Scholar]
- 21.Zagulska-Szymczak, S., Filipkowski, R. K. & Kaczmarek, L. (2001) Neurochem. Int. 38, 485-501. [DOI] [PubMed] [Google Scholar]
- 22.Gingras, A. C., Raught, B. & Sonenberg, N. (2001) Genes Dev. 15, 807-826. [DOI] [PubMed] [Google Scholar]
- 23.Wisden, W. & Morris, B. J. (1994) in In Situ Hybridization Protocols for the Brain, eds. Wisden, W. & Morris, B. J. (Academic, San Diego), pp. 9-34.
- 24.Kennelly, P. J. & Krebs, E. G. (1991) J. Biol. Chem. 266, 15555-15558. [PubMed] [Google Scholar]
- 25.Kemp, B. E. & Pearson, R. B. (1990) Trends Biochem. Sci. 15, 342-346. [DOI] [PubMed] [Google Scholar]
- 26.Pinna, L. A. & Ruzzene, M. (1996) Biochim. Biophys. Acta 1314, 191-225. [DOI] [PubMed] [Google Scholar]
- 27.French, P. J., O'Connor, V., Jones, M. W., Davis, S., Errington, M. L., Voss, K., Truchet, B., Wotjak, C., Stean, T., Doyere, V., et al. (2001) Eur. J. Neurosci. 13, 968-976. [DOI] [PubMed] [Google Scholar]
- 28.Bliss, T. V. & Lomo, T. (1973) J. Physiol. 232, 331-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang, S. J., Reis, G., Kang, H., Gingras, A. C., Sonenberg, N. & Schuman, E. M. (2002) Proc. Natl. Acad. Sci. USA 99, 467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan, L., Chang, J. S., Costa, A. & Schedl, P. (2001) Development (Cambridge, U.K.) 128, 1159-1169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.