Abstract
In yeasts and mammals, PEX10 encodes an integral membrane protein with a C3HC4 RING finger motif in its C-terminal domain and is required for peroxisome biogenesis and matrix protein import. In humans, its dysfunction in peroxisome biogenesis leads to severe Zellweger Syndrome and infantile Refsum disease. Here we show that dysfunction of a homologous gene in Arabidopsis leads to lethality at the heart stage of embryogenesis, impairing the biogenesis of peroxisomes, lipid bodies, and protein bodies. In a T-DNA insertion mutant disrupting the fourth exon of the AthPEX10 gene, ultrastructural analyses fail to detect peroxisomes characteristic for wild-type embryogenesis. Storage triacyl glycerides are not assembled into lipid bodies (oil bodies; oleosomes) surrounded by the phospholipid–protein monolayer membrane. Instead, the dysfunctional monolayer membranes, which derive from the bilayer membrane of the endoplasmic reticulum, accumulate in the cytosol. Concomitantly the transfer of the storage proteins from their site of synthesis at the endoplasmic reticulum to the vacuoles is disturbed. The mutant can be rescued by transformation with wild-type AthPEX10 cDNA. Transformants of wild-type Hansenula polymorpha cells with the AthPEX10 cDNA did produce the encoded protein without targeting it to peroxisomes. Additionally, the cDNA could not complement a Hansenula pex10 mutant unable to form peroxisomes. The ultrastructural knockout phenotype of AthPEX10p suggests that this protein in Arabidopsis is essential for peroxisome, oleosome, and protein transport vesicle formation.
Availability of DNA sequences from partial or complete genomes has led to prediction of functions of genes or members of gene families across different species. Conserved coding domains characteristic for a gene with known biochemical, cell biological, or structural function in one or several organisms are used for these predictions, and the gene is named accordingly. Subsequent analysis of mutants with codon changes, deletions, or knockout insertions reveals the gene to have different or additional functions. Examples are provided among the 25 peroxisome biogenesis (PEX) genes of the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Yarrowia lipolytica and Pichia pastoris, of Homo sapiens, and of Arabidopsis thaliana (1–3). Peroxins function in early peroxisome biogenesis (PEX1, PEX6), peroxisome membrane protein import (PEX3, PEX16, PEX19), targeting receptors (PEX5, PEX7), matrix protein import (PEX2, PEX10, PEX12, PEX13, PEX14, PEX17), receptor recycling (PEX1, PEX4, PEX6), and proliferation (PEX11).
PEX10, PEX12, and PEX2 encode integral membrane proteins mutually distinct in the primary sequence except for a shared C3HC4-type Zn-binding RING finger motif in their C-terminal domain. In mammals and yeasts, they have been shown to interact with each other; impaired function of these three peroxins results in the failure of matrix protein import, implying that RING peroxins are involved in the process of protein transport across the membrane (4–6). The TLGEEY-motif present in the N-terminal part of PEX10p is typical for this subfamily of Zn-binding proteins (7).
In P. pastoris, the loss of PEX10p does not interfere with the formation of peroxisome membranes or peroxisome proliferation. However, PEX10p is required for import of matrix proteins containing either type of peroxisomal targeting signal as well as for the elaboration of the peroxisome lumen; loss of PEX10p leads to accumulation of membrane sheets (8, 9). In H. polymorpha, PEX10-deficient mutants contain no peroxisome-like structures, and peroxisomal enzymes are located in the cytosol, whereas overexpression of PEX10p leads to increased numbers of peroxisomes, indicating its role in peroxisome proliferation (10). PEX10-deficient cells in humans show peroxisome shells that import membrane proteins, but no matrix proteins. Therefore, targeting of human PEX10p to peroxisome membranes can be uncoupled from the matrix protein import machinery (4, 7, 11). Loss of PEX10p leads to Zellweger syndrome, a severe form of the peroxisome biogenesis disorder (5). The above studies reveal that presence of the Zn-finger and the TLGEEY domain does not necessarily identify PEX10 peroxins (PEX10p) with identical functions.
PEX12p-deficient cells fail to import peroxisomal matrix proteins through both the PTS1 and PTS2 pathway in P. pastoris, S. cerevisiae, and human (4, 12, 13). These cells retain particular structures that contain peroxisomal membrane proteins (“ghosts”), indicating that mutant pex12 cells are not impaired in peroxisomal membrane biogenesis.
Mutations in the PEX2 gene in Y. lipolytica lead to the accumulation of three peroxisomal subpopulations and prevent the formation of mature functional peroxisomes (14, 15). The irregular vesicular structures are surrounded by multiple unit membranes; some peroxisomes in PEX2 mutants are associated with elements of the endoplasmic reticulum (ER) and Golgi. The subpopulations may represent intermediates in peroxisome assembly. Homozygous PEX2-deficient mice survive in utero but die several hours after birth (16). The mutant animals lack normal peroxisomes but assemble empty membrane ghosts and display abnormal peroxisomal biochemistry.
The AthPEX2 gene encoding a C3HC4-type RING finger protein with two putative membrane-spanning domains has originally been cloned as TED3 (17). Mutant det3 functions as a dominant suppressor of det1 mutants with aberrant photomorphogenesis. T-DNA insertions in the seventh intron or eighth exon of the gene yielded T2 and T3 plants heterozygous for the insertion but no homozygous plants, suggesting embryonic lethality. Interactions of TED3 with DET1 and COP1 and their suppressor mutants were analyzed with regard to photomorphogenesis of Arabidopsis. These results reveal that peroxines are proteins with multiple functions.
We have cloned an Arabidopsis gene encoding a protein with a C3HC4-type RING finger and a TLGEEY sequence and designated it as AthPEX10 (18). Here we report that loss of function of this protein in a T-DNA insertion mutant leads to embryo lethality. Formation of lipid bodies, protein bodies, and peroxisomes are aborted at the heart stage of the developing Arabidopsis embryo, and ER-derived membrane stacks accumulate in the cytosol.
Materials and Methods
Isolation of Two A. thaliana PEX10 Transposon Insertion Lines. The catalog of the Nottingham Arabidopsis Stock Center contained a PEX10 knockout line (ecotype Landsberg erecta, accession no gi:2739359, code N100156) carrying a single copy of a modified maize transposable dissociation element (DsG) (19) inserted within exon 4 of PEX10 at nucleotide 631 (for further detail, see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org). Two lines of the obtained seeds showed segregation for the transgene on Murashige and Skoog plates containing 50 mg/liter kanamycin.
Transformation of Mutant A. thaliana with the AthPEX10 cDNA. PEX10-cDNA (18) was amplified with the sense primer (5′-AAC TAG CTA GCA TGA GGC TTA ATG GGG ATT CGG-3′), which contains a NheI restriction site near the start methionine codon, and the antisense primer (5′-TTG AGC TCG TCT TGT AAT AGA TCT CCA AT-3′), which introduces a SacI restriction site in the 3′ UTR of PEX10-cDNA just upstream of the poly(A)-tail. The 1,493-bp PCR product was cloned between the constitutive 35S S1 promoter and the nos transcriptional terminator into binary vector pBI121 (CLONTECH), carrying a Bar resistance gene. Transformation of Agrobacterium tumefaciens strain C58 with the Ti-plasmid pGV3850 was achieved by electroporation, and transformation of the transgenic A. thaliana lines DsG-3 and DsG-7 segregating for the insertion in Pex10 was performed by floral dipping (20).
Electron Microscopy. Embryos were fixed for 2 h at room temperature with 2.5% glutaraldehyde in fixative buffer (75 mmol/liter cacodylate/2 mmol/liter MgCl2, pH 7.0). After the cells had been rinsed several times with the same buffer, they were postfixed for 2 h with 2% osmium tetroxide in fixative buffer. After washing steps with fixative buffer and distilled water the embryos were dehydrated in a graded series of acetone solution: 10%, 20%, 40%, 60%, 80%, and 100% (en bloc staining was carried out with 1% uranylatetate at the 20% stage). Embryos were infiltrated and embedded in Spurr's low-viscosity resin. Ultrathin sections were examined in a Zeiss EM 912 transmission electron microscope equipped with an integrated OMEGA energy filter, operated at 80 kV in the zero loss mode.
Results
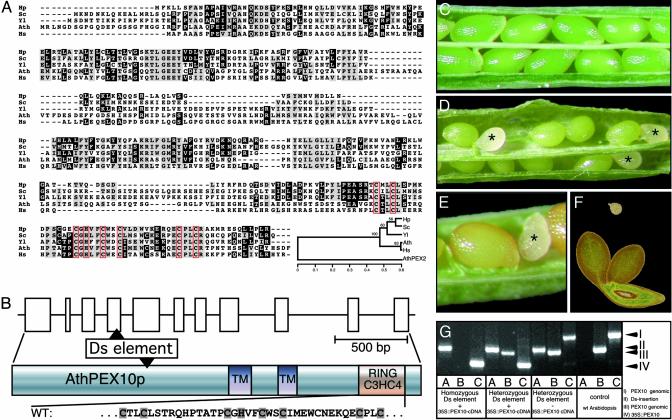
The Arabidopsis PEX10 Gene. The A. thaliana PEX10 is a singlecopy gene with 11 exons encoding a 43-kDa protein with 381 aa (18). The predicted AthPEX10p is 35% identical and 56% similar to the human, H. polymorpha, and Y. lipolytica homologues; 30% identity and 50% similarity is found to the S. cerevisiae homologue; the evolutionary tree was calculated on the basis of the alignment of the Zn-finger regions with AthPEX2 as an outgroup (Fig. 1A). Two putative membrane-spanning domains are in exons 6 and 8, respectively, whereas the RING finger motif spans exons 10 and 11 (Fig. 1B).
Fig. 1.
The A. thaliana PEX10 gene and its seed lethal T-DNA disruption phenotype. (A) Comparison of the amino acid sequences of PEX10 peroxins. Hp, H. polymorpha; Sc, S. cerevisiae; Yl, Y. lipolytica; Ath, A. thaliana; Hs, Homo sapiens. The TLGEF-motif and the two transmembrane helices are shaded. The amino acids of the Zn-binding C3HC4 Ring finger are framed. The family tree has been constructed with the neighbor-joining algorithm with AthPEX2 as an outgroup. (B) Exon–intron structure of the AthPEX10 gene with the DsG element insertion. (C) Wild-type silique with uniformly maturing green seeds with green embryos. (D) White lethal seeds (asterisk) in the silique from a heterozygous T2 plant segregating for the disrupted PEX10 gene. (E) Brown mature and white lethal seed. (F) Lethal embryo at the heart stage (above) and viable torpedo-stage embryo (below). (G) Identification by PCR of all possible genotypes in T2 segregants of plants homozygous for the DsG disrupted PEX10 and rescued with a PEX10-cDNA.
Phenotype of a PEX10 Knockout Mutant and Its Rescue by Transformation with the PEX10 cDNA. A T-DNA insertion line carrying a Ds transposon element and the NPTII gene, which confers kanamycin resistance, was obtained from The National University of Singapore (19). The insertion disrupted the AthPEX10p after amino acid 125 encoded in the fourth exon (Fig. 1B). Segregation of T2 seedlings on kanamycin-containing media gave 63.9% resistant plants instead of the expected 75%, indicating lethality of the homozygous insertion mutants. This finding was verified by dissecting individual maturing siliques (Fig. 1 C–F). Wild-type siliques showed consistently maturing seeds with green embryos of uniform shape and size (Fig. 1C); unpollinated ovules are found occasionally. In young siliques of the plants segregating kanamycin-resistant seedlings, chlorophyll-free, immature, lethal seeds were present beside normally maturing green seeds (Fig. 1D). The white seeds stagnated in their development, whereas the viable, green seeds developed into brown mature seeds (Fig. 1E). Analyses of two heterozygous lines yielded a frequency of 21.3% ± 7.1% (SD) and 21.6% ± 4.5% lethal seeds in agreement with a recessive lethal segregation of the embryos homozygous for the insertion. In a sample of 103 lethal embryos, 29% arrested at the late globular stage, whereas 68% died at the heart stage, and only 3% reached the torpedo stage (Fig. 1F and Fig. 4, which is published as supporting information on the PNAS web site).
To prove that seed lethality was caused by the dysfunctional PEX10 gene, we investigated whether the T-DNA insertion line could be rescued by transformation with the PEX10-cDNA. Transgenic Arabidopsis plants heterozygous for the Ds element insertion in PEX10 were transformed with the PEX10-cDNA under the control of the 35S S1-promoter together with a bar gene providing resistance to bialaphos for selection of the transformants. Twenty T1 plants (the primary transgenic plants) viable on kanamycin and bialaphos were obtained. Their genomic DNA was isolated from rosette leaves and analyzed by PCR to confirm hemizygosis for the Ds transposon in the PEX10 gene and presence of the PEX10-cDNA.
For identification of plants homozygous for the Ds disrupted PEX10 gene and the complementing PEX10 cDNA, three pairs of PCR primers were designed that permitted genotyping of all possible T2 segregants. A sense primer annealing at the start codon of the PEX10 gene 5′-ATG AGG CTT AAT GGG GAT TCG-3′ amplifies with the antisense primer 5′-CAC AGG CCA TCG CTG AAT AG-3′ in the fifth exon a 922-bp PCR product diagnostic for the presence of at least one intact genomic copy of PEX10 (Fig. 1G Right, lane C, PCR band I), whereas a 588-bp product is amplified from the transgenic PEX10-cDNA (Fig. 1G Left, lane C, PCR band IV). Because amplification of the longer genomic fragment is inefficient in the presence of the cDNA transgene, an antisense primer in the fourth intron 5′-GAC ATT TAT ATC AAT ATC AGC GG-3′ that specifically amplifies a 727-bp genomic PEX10 fragment was used to detect the endogenous PEX10 gene in the presence of the cDNA transgene (Fig. 1G Center and Right, lanes B, PCR band III). The Ds element insert in PEX10 was detected with the third antisense primer 5′-GGT TCC CGT CCG ATT TCG ACT-3′ annealing to position 6733–53 of the Ds element in the fourth exon and yielding an 809-bp DNA fragment (Fig. 1G Left and Center, lanes A, PCR band II). When we used this strategy, we found plants homozygous for the Ds element that were complemented by transformation with the PEX10 cDNA (Fig. 1G Left) as well as plants heterozygous for the Ds element carrying the PEX10-cDNA (Fig. 1G Middle Left) and plants heterozygous for the Ds element without the PEX10 cDNA (Fig. 1G Middle Right) in the expected ratio. A. thaliana wild-type plants served as a negative control (Fig. 1G Right). Expression of PEX10 cDNA in transgenic pex10 plants thus complements the embryo lethality, making the plants indistinguishable from wild-type plants. The size of the fragment, 809 bp amplifiable with the sense primer and the antisense primer annealing at the 3′ end of the gene trap element DsG, reveals that DsG is inserted with its 3′ end in the reading frame direction of the Pex10 gene.
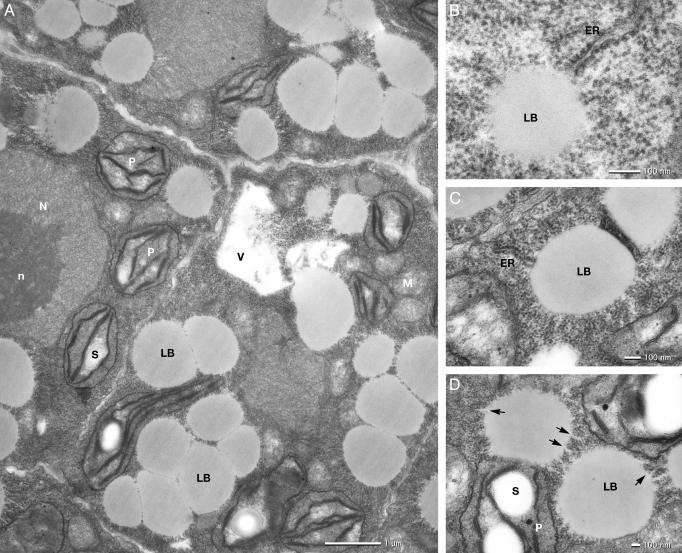
Ultrastructure of Normally Developing Embryos Subsequent to the Stages at Which the T-DNA Insertion Mutant Embryos Derail. Most of the embryos with the nonfunctional PEX10p died at the heart stage. Embryo development within a silique is highly synchronous. The lethal embryos could therefore be identified as being at an apparently younger stage than the rest of the embryos. They were analyzed by electron microscopy. The wild-type heart stage embryo contains meristematic type cells with developing primary cell walls (Fig. 5, which is published as supporting information on the PNAS web site). The ER is well developed and studded with ribosomes (rough ER). The cytosol is densely packed with ribosomes. Profiles of well developed Golgi stacks, mitochondria, peroxisomes, and developing chloroplasts with grana stacks of two to four appressed thylakoid disks are seen in the electron micrographs. The cells contain one or several vacuoles (21). In the torpedo stage, the general cell and organelle structures are the same as in the heart stage. However, a highly active synthesis of lipid bodies is in progress in the cytosol with its densely packed ribosomes (Fig. 2A). On the other hand, only an occasional vacuole with indication of storage protein deposition is seen. The triacylglycerols to be deposited in the lipid bodies of A. thaliana comprise saturated and unsaturated fatty acids synthesized in the chloroplast, transfered into an acyl–CoA pool in the ER, and converted to di- and triacylglycerols in the membranes of the ER (22). At the same time, oleosin proteins are synthesized on the cytosolic side of the ER and inserted into the outer phospholipid layer of the membrane (23). Ultrastructural evidence supports the view that the triacylglycerols accumulate between the inner and outer leaflet of the ER membrane and that the single membrane of the lipid body thus arises from the phospholipidlayer with the inserted oleosins (ref. 24 and Fig. 2 B and C). The developing lipid bodies are characterized by a surface of regular spaced channels with a diameter of 10 nm (Fig. 2D). In high-resolution transmission electron micrographs, these channels can be seen to have the same electron density as the triacylglycerols in the lipid body and are directly connected to the profiles of the ER.
Fig. 2.
Electron micrographs of a ultrathin section through a wild-type torpedo-stage embryo. Electron translucent lipid bodies (LB) are formed in the cytoplasm, which is tightly packed with ribosomes (A). Rough ER is in connection with spherical lipid bodies (B and C). The developing lipid bodies are characterized by a surface of regular spaced channels (D, arrows) with the same electron density as the triacylglycerols of the lipid bodies. N, nucleus; n, nucleolus; P, chloroplast; S, starch grain; V, vacuole.
We counted the number of peroxisome profiles visible in the heart and torpedo-stage embryo, and found 4.3 peroxisomes per 100 μm2 cytoplasmic area. There was no obvious difference among the different developmental stages.
During the further development of the embryo (walking stick stage and cotyledon differentiation), secondary cell walls are formed and the storage protein and the globoid crystals are synthesized and deposited in large vacuoles (ref. 25 and Fig. 6, which is published as supporting information on the PNAS web site). The lipid bodies are pressed against the plasma membrane, and well developed chloroplasts are positioned around the centrally located protein storage bodies and the nucleus, mitochondria, peroxisomes, and short profiles of the ER. During the final maturation of the embryo, the cytosol is concentrated around the nucleus and a few spaces between lipid body clusters. The thylakoids of the chloroplasts have been eliminated and the plastid has been reduced to small proplastids with stroma and a few plastoglobuli. Prominent short profiles of the ER, mitochondria, and ricinosomes are distinguishable. Ricinosomes are identified by their single surrounding membrane studded with ribosomes (26).
Ultrastructural Features of the Lethal Embryo Pathology. The lethal embryos reach only the size of the heart stage embryo. Their cellular differentiation at the time of cell death is a mosaic of mature embryo differentiation and an arrested development of lipid bodies and protein bodies. The cells have formed secondary cell walls. Vacuoles of the size similar to wild-type are present; the content, however, is limited to uncompacted material (Fig. 7, which is published as supporting information on the PNAS web site). Characteristic peroxisomes were not observed. Densely packed lipid bodies of the size and shape typical for the wild-type are absent. The cytosol contains mitochondria and numerous vesicles, as well as dedifferentiated proplastids like in wild-type, but containing larger starch granules. Extended profiles of the ER are found as well as the short profiles typical of the wild-type mature embryo.
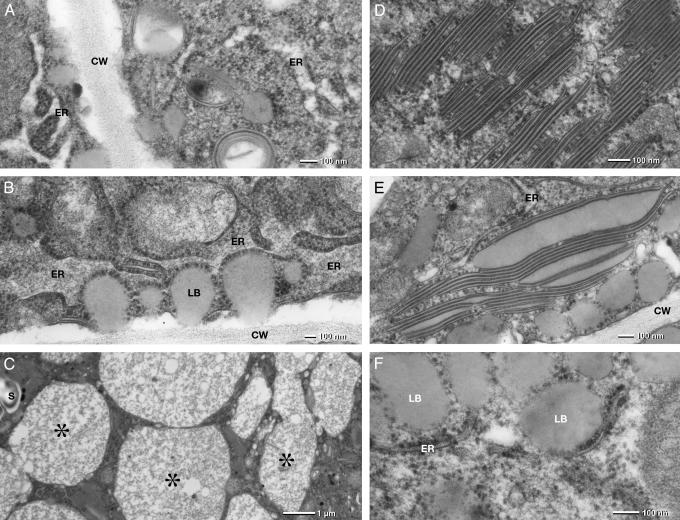
There are three abnormal structures that give a hint for the role of PEX10p in the filling of the storage vacuoles with storage proteins and the formation of lipid bodies. In the wild-type, albumins and globulins are synthesized by the polysomes of the ER, transferred into its lumen, and then transported by vesicles from the ER via the Golgi apparatus into the storage vacuoles. In the lethal cells, the rough ER forms an extended network with swollen ER lumen (Fig. 3A). The swollen ER becomes more and more elaborated (Fig. 3B) and forms numerous vacuoles similar in size and shape of wild-type protein bodies. In contrast to wild-type protein bodies, they only contain amorphous, fibrous protein material (Fig. 3C), which obviously tends to aggregate (Fig. 7, asterisk) and form electron dense precipitates (Fig. 3C, asterisk). The absence of PEX10p thus might prevent the budding of the vesicles transporting the protein from the ER to the Golgi and from there to the vacuoles.
Fig. 3.
Electron micrographs of a ultrathin section through the meristematic cells of a lethal embryo. The rough ER forms an elaborate network of “swollen” strands and cisternae (A). The electron translucent lumen of the ER becomes filled with amorphous, fibrous material (B), which finally fills storage vacuoles of similar size as wild-type protein bodies (C, asterisks). Abnormal lipid body formation is a characteristic feature: extemly thin “lipid body disks” form stacks of several disks (D). The flat “lipid bodies” have a tripartite structure: two electron-dense half-unit membranes and a homogeneous, electron-translucent matrix presumably formed by triacylglycerols. The thickness of the lipid matrix of the flat disks is in the range of 10 nm but can differ widely (E). Little lipid bodies (LB) can apparently be assembled by the ER in the absence of PEX10p and are reminiscent of nascent lipid bodies (oleosomes) found at the torpedo stage (B and F). CW, cell wall.
The second abnormal structure consists of stacks of thin, highly compressed membrane leaflets in parallel or spheroidal arrangement (Fig. 3 D and E). A single leaflet is characterized by a bordering electron dense half-unit membrane and a homogenous less electron dense matrix obviously representing storage lipid. The thickness of the lipid matrix varies (Fig. 3E), but is typically in the range of 10 nm (Fig. 3D). Because the included material has the same electron density as the lipid bodies, we speculate that the leaflets are extremely flat lipid bodies. Conceivably then, the leaflets represent the initial form of the lipid bodies and the space between dark lines would be filled with triacylglycerols. PEX10p appears to be required for the formation of lipid bodies from the half-unit membranes that organize the accumulation of triacylglycerides.
The third abnormal structures are small lipid bodies with a half-unit membrane studded with ribosomes (Fig. 3 B and F). This type of lipid bodies can apparently be assembled by the ER in the absence of PEX10p and is reminiscent of nascent lipid bodies found at the torpedo stage in A. thaliana (21) and in maturing Cucurbita and Citrullus cotyledons (24).
AthPEX10p Cannot Complement a H. polymorpha pex10-Deficient Mutant Lacking Peroxisomes. Transformants expressing AthPEX10p with the alcohol oxidase promoter in a pex10 mutant H. polymorpha (10) did not allow the cells to grow on methanol; morphologically recognizable peroxisomes were not induced. Transformants overexpressing AthPEX10p in wild-type H. polymorpha did not target the protein into the peroxisomes of the cell. On differential centrifugation of cell extracts, AthPEX10p was found in the 30,000 × g supernatant together with cytosolic proteins, small membranous structures, and matrix proteins of broken organelles, but not in the pellet with peroxisomes and mitochondria. In an analysis of a sucrose gradient from homogenized, methanol-grown AthPEX10p::wtHp cells, the peroxisomal enzyme alcohol oxidase peaked at a sucrose concentration of 57–50%, the mitochondrial enzyme cytochrome c oxidase at a concentration of 45–35%, similar to untransformed wild-type cells. AthPEX10p, however, was exclusively found in the upper fractions 18–23 of the gradient together with alcohol oxidase from broken peroxisomes (see Supporting Text). We conclude that the membrane-targeting signal of the plant PEX10p is probably different from that of H. polymorpha.
Discussion
The mechanism of peroxisome biogenesis is still obscure. Because membranes are not considered to be formed de novo, peroxisomes have to arise by either one of two pathways: one would be PEX11-mediated division and proliferation of preexisting peroxisomes; the other pathway would originate from intracellular membranes such as the ER.
In the yeast Y. lipolytica (27–29), PEX2p and PEX16p are sorted via the ER to the peroxisomes, where they are localized as integral and peripheral membrane proteins, respectively. Mutants in PEX1, PEX2, PEX6, and PEX9 compromise not only peroxisome biogenesis but also affect protein secretion and prevent exit of extracellular proteases from the ER. In H. polymorpha, the membrane-bound peroxin PEX3p accumulated in the ER both when overproduced and when cells were incubated with Brefeldin, a toxin that prevents the formation of ER-derived vesicles (30, 31).
In plants, to date only AthPEX14p has been localized to the peroxisomal membrane (32). Two pathways for sorting of peroxisomal membrane proteins (PMP) have been proposed for plants: direct sorting is deduced from in vitro insertion of Arabidopsis PMP22 into isolated sunflower peroxisomes; or indirect sorting via the ER from studies with ascorbate peroxidase, a constitutive component of oilseed seedling glyoxysomes and leaf peroxisomes (3).
Localization of PEX16p varies with species. In Y. lipolytica, it is a peripheral membrane protein associated with the matrix side of peroxisomes (29), whereas in humans it behaves as an integral membrane protein with both of its termini exposed to the cytoplasm. Inactivation of the human PEX16 abolishes the synthesis of peroxisomes; expression of PEX16 restores the formation of new peroxisomes in the absence of preexisting peroxisomes. Human PEX16p is detected at steady state synthesis only in peroxisomes, and no other compartment. Brefeldin did not inhibit the targeting of PEX16myc to peroxisomes (33).
The recessive A. thaliana pex16 T-DNA insertion mutant was originally isolated as a shrunken seed mutant sse1 that lacks storage protein bodies in the mature grain and contains few lipid bodies; instead, starch granules accumulate and the seeds cannot germinate. The cloned gene can fully complement the deficient seed development (34). AthPEX16p has 26% identity to PEX16p of Y. lipolitica with similar arrangements of hydrophobic and hydrophilic domains (14) and complements pex16 mutants defective in the formation of peroxisomes and the transportation of plasma membrane and cell wall-associated proteins. Lin and coworkers (34) examined dry seeds, where peroxisomes are not generally found, and thus the involvement of AthPEX16 in peroxisome formation in A. thaliana remains to be demonstrated.
Our cytological obser vations with the dysfunctional AthPEX10p indicate that it is directly or indirectly involved in the formation of peroxisomes, lipid bodies, and protein bodies, thus influencing the formation of ER-derived vesicles. Our data suggest that AthPEX10p is sorted to peroxisomes or import-competent peroxisome primordia via the ER. Four distinct secretory pathways serve protein secretion, cell surface growth, and peroxisome biogenesis in Y. lipolytica; at least two of these secretory pathways, which are involved in the export of proteins to the external medium and in the delivery of proteins for assembly of the peroxisomal membrane, diverge at the level of the ER (33). By analogy, AthPEX10p could participate in peroxisome and storage organelle formation. The vesicles and stacks of membranes in the Arabidopsis pex10 mutant resemble subcellular structures in the Y. lipolytica pex10 mutant.
Taken together, the results reveal that the function of PEX10p and other peroxins is not limited to peroxisome biogenesis. Peroxins play multiple roles in the formation of various organelle membranes derived from the single or double phospholipid layer of the ER membrane.
Supplementary Material
Abbreviation: ER, endoplasmic reticulum.
References
- 1.Titorenko, V. L. & Rachubinski, R. A. (2001) Trends Cell Biol. 11, 22-29. [DOI] [PubMed] [Google Scholar]
- 2.Charlton, W. & Lopez-Huertas, E. (2002) in Plant Peroxisomes, eds. Baker, A. & Graham, I. A. (Kluwer Academic, Dordrecht, The Netherlands), pp. 385-426.
- 3.Mullen, R. T., Flynn, C. R. & Trelease, R. N. (2001) Trends Plant Sci. 6, 256-261. [DOI] [PubMed] [Google Scholar]
- 4.Chang, C. C., Warren, D. S., Sacksteder, K. A. & Gould, S. J. (1999) J. Cell Biol. 147, 761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okumoto, K., Itoh, R., Shimozawa, N., Suzuki, Y., Tamura, S., Kondo, N. & Fujiki, Y. (1998) Hum. Mol. Genet. 7, 1399-1405. [DOI] [PubMed] [Google Scholar]
- 6.Okumoto, K., Abe, I. & Fujiki, Y. (2000) J. Cell Biol. 275, 25700-25710. [DOI] [PubMed] [Google Scholar]
- 7.Warren, D. S., Morrell, J. C., Moser, H. W., Valle, D. & Gould, S. J. (1998) Am. J. Hum. Genet. 63, 347-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalish, J. E., Theda, C., Morrell, J. C., Berg, J. M. & Gould, S. J. (1995) Mol. Cell. Biol. 15, 6406-6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder, W. B., Faber, K. N., Wenzel, T. J., Koller, A., Luers, G. H., Rangell, L., Keller, G. A. & Subramani, S. (1999) Mol. Biol. Cell 10, 1745-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan, X., Waterham, H. R., Veenhuis, M. & Cregg, J. M. (1995) J. Cell Biol. 128, 307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren, D. S., Wolfe, B. D. & Gould, S. J. (2000) Hum. Mutat. 15, 509-521. [DOI] [PubMed] [Google Scholar]
- 12.Kalish, J. E., Keller, G. A., Morell, J. C., Mihalik, S. J., Smith, B., Cregg, J. M. & Gould, S. J. (1996) EMBO J. 15, 3275-3285. [PMC free article] [PubMed] [Google Scholar]
- 13.Albertini, A. M., Girzalsky, W., Veenhuis, M. & Kunau, W. H. (2001) Eur. J. Cell Biol. 80, 257-270. [DOI] [PubMed] [Google Scholar]
- 14.Eitzen, G. A., Titorenko, V. I., Smith. J. J., Veenhuis, M., Szilard, R. K. & Rachubinski, R. A. (1996) J. Biol. Chem. 271, 20300-20306. [DOI] [PubMed] [Google Scholar]
- 15.Titorenko, V. I., Eitzen, G. A. & Rachubinski, R. A. (1996) J. Biol. Chem. 34, 20307-20314. [DOI] [PubMed] [Google Scholar]
- 16.Faust, P. L., Su, H. M., Moser, A. & Moser, H. W. (2001) J. Mol. Neurosci. 16, 289-297. [DOI] [PubMed] [Google Scholar]
- 17.Hu, J., Aguirre, M., Peto, C., Alonso, J., Ecker, J. & Chory, J. (2002) Science 297, 405-409. [DOI] [PubMed] [Google Scholar]
- 18.Schumann, U., Gietl, C. & Schmid, M. (1999) Plant Physiol. 119, 1147. [Google Scholar]
- 19.Parinov, S., Sevugan, M., Ye, D., Yang, W.-C., Kumaran, M. & Sundaresan, V. (1999) Plant Cell 11, 2263-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer, K., Leube, M. P. & Grill, E. (1994) Science 264, 1452-1455. [DOI] [PubMed] [Google Scholar]
- 21.Bowman, J. (1994) Arabidopsis: An Atlas of Morphology and Development (Springer, New York).
- 22.Somerville, C., Browse, J., Jaworski, J. G. & Ohlrogge, J. B. (2000) in Biochemistry and Molecular Biology of Plants, eds. Buchanan, B. B., Gruissem, W. & Jones, R. L. (Am. Soc. Plant Physiol., Rockville, MD), pp. 456-527.
- 23.Huang, A. H. C. (1996) Plant Physiol. 110, 1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wanner, G., Formanek, H. & Theimer, R. R. (1981) Planta 151, 109-123. [DOI] [PubMed] [Google Scholar]
- 25.Otegui, M. S., Capp, R. & Staehelin, L. A. (2002) Plant Cell 14, 1311-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gietl, C. & Schmid, M. (2001) Naturwissenschaften 88, 49-58. [DOI] [PubMed] [Google Scholar]
- 27.Titorenko, V. I. & Rachubinski, R. A. (1998) Mol. Cell. Biol. 18, 2789-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eitzen, G. A., Szilard, R. K. & Rachubinski, R. A. (1997) J. Cell Biol. 137, 1265-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Titorenko, V. I., Ogrydziak, D. M. & Rachubinski, R. A. (1997) Mol. Cell. Biol. 17, 5210-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baerends, R. J. S., Rasmussen, R. E., Hilbrands, R. E., van der Heide, M., Faber, K. N., Reuvekamp, P. T. W., Kiel, J. A. K. W., Cregg, J. M., van der Klei, I. J. & Veenhuis, M. (1996) J. Biol. Chem. 271, 8887-8894. [DOI] [PubMed] [Google Scholar]
- 31.Salomons, F. A., van der Klei, I. J., Kram, A. M., Harder, W. & Veenhuis, M. (1997) FEBS Lett. 411, 133-139. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi, M., Nito, K., Toriyama-Kato, K., Kondo, M., Yamaya, T. & Nishimura, M. (2000) EMBO J. 19, 5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.South, S. T. & Gould, S. J. (1999) J. Cell Biol. 144, 255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, Y., Sun, L., Nguyen, L. V., Rachubinski, R. A. & Goodman, H. M. (1999) Science 284, 328-330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.