Abstract
Gene silencing mediated by double-stranded RNA is a sequence-specific RNA degradation mechanism highly conserved in eukaryotes that serves as an antiviral defense pathway in both plants and Drosophila. Short interfering RNAs (siRNAs), the 21- to 23-nt double-stranded intermediates of this natural defense mechanism, are becoming powerful tools for reducing gene expression and countering viral infection in a variety of mammalian cells. Here we report the use of siRNAs to target reporter gene expression and viral DNA accumulation in cultured plant cells. Transient expression of reporter genes encoding either GFP or red fluorescent protein from Discosoma was specifically reduced by 58% and 47%, respectively, at 24 h after codelivery of cognate siRNAs in BY2 protoplasts. In contrast to mammalian systems, the siRNA-induced silencing of GFP expression was transitive as indicated by the presence of siRNAs representing parts of the target RNA outside the region homologous to the triggering siRNA. Codelivery of an siRNA designed to target the mRNA encoding the replication-associated protein (AC1) of the geminivirus African cassava mosaic virus (ACMV) from Cameroon blocked AC1 mRNA accumulation by ≈91% and inhibited accumulation of the ACMV genomic DNA by ≈66% at 36 and 48 h after transfection. As with siRNA-induced reporter gene silencing, the siRNA targeting ACMV AC1 was specific and did not affect the replication of East African cassava mosaic Cameroon virus. This report demonstrates the occurrence of siRNA-mediated suppression of gene expression in cultured plant cells and that siRNA can interfere with and suppress accumulation of a nuclear-replicated DNA virus.
Posttranscriptional gene silencing (PTGS) is a sequence-specific defense mechanism that can target both cellular and viral mRNAs for degradation and is widely used as a tool for inactivating gene expression (1, 2). PTGS was discovered in plants (3), and a closely related phenomenon, RNA interference, is known to occur in a wide range of organisms including Caenorhabditis elegans, Neurospora crassa, Drosophila melanogaster, and mammals (4–7). Transgenes and viruses can induce gene silencing in plants, and it was proposed that PTGS is a natural defense mechanism against virus accumulation (8). The process is initiated by double-stranded RNA (dsRNA) molecules, possibly generated by replicative intermediates of viral RNAs or by aberrant RNAs, which become dsRNA by host-encoded RNA-dependent RNA polymerase activity (9–11). These dsRNAs are cleaved by Dicer-like enzymes into short interfering RNAs (siRNAs) of between 21 and 26 nt (12) in length, which then promote RNA degradation by forming a multicomponent RNA-induced silencing complex that destroys cognate mRNA (13–15).
Virus-induced gene silencing has been demonstrated for a number of RNA and DNA viruses (2, 16, 17), with the production of virus-specific siRNAs in the case of potato virus X-infected plants (8). As a counterdefense strategy, certain viruses encode suppressor proteins that can act at different points in the RNA-silencing pathway (2, 18, 19). Vectors based on two geminiviruses that carried host-related inserts, tomato golden mosaic virus and cabbage leaf curl virus, silenced the homologous genes in Nicotiana benthamiana and Arabidopsis thaliana, respectively (20–22). Although AC2 of the African cassava mosaic virus (ACMV) from Kenya (19) and C2 of tomato yellow leaf curl China virus (23) have been identified as mild viral suppressors of PTGS, the exact mode of action of AC2 in the silencing pathway is not clear. In our laboratory, we observed that certain geminiviruses are capable of inducing PTGS with the production of virus-specific siRNAs, the hallmark of PTGS, in N. benthamiana and in cassava plants, resulting in recovery phenotype (unpublished data) and prompting us to question whether gene silencing can be used as a tool to counter geminivirus infection. Geminiviruses infect a wide variety of economically important crops worldwide (24, 25). These viruses have a single-stranded DNA (ssDNA) genome that is replicated in the nuclei of infected plant cells by a rolling-circle mechanism (26). Among the different gene products encoded by the virus, only AC1, the replication-associated protein, is indispensable for viral DNA replication (26, 27). Therefore, we envisaged the usage of siRNA, an intermediate in the silencing pathway, to target the AC1 gene, which would be a valuable tool to counter geminiviruses. This would also shed insight into the function of suppressors (AC2) and whether they could block siRNAs to prevent gene silencing.
dsRNA is remarkably effective at suppressing specific gene expression in a number of organisms including plants. Recently it was shown that siRNAs of 21 nt in size, an intermediate of the RNA-interference pathway, are effective in suppression of gene expression in animal systems including mammals. Gene silencing induced by long dsRNA constructs has been demonstrated in transgenic plants showing resistance to virus infection (9): transient interference with endogenous gene expression (28), with virus infection in plants (29), and with reporter gene expression in plant cells (30, 31). The siRNA technology offers animal virologists a new way to control viruses because the use of long dsRNA triggers the IFN response, which leads to apoptosis in most mammalian cells (32). Introduction of siRNA into mammalian cells can suppress expression of specific endogenous genes and can target a number of viruses (7, 33, 34). These observations have led to the use of siRNAs as a tool for gene therapy (34–36). Although the potato virus X vector carrying a short length of RNA was capable of inducing PTGS, the possibility that viral dsRNA by itself was the PTGS inducer was not ruled out (37). In this report we demonstrate that the use of siRNAs is very effective in isolated plant cells. Protoplasts provide a method for rapid and quantitative analysis of transgene expression before committing to the process of transgenic plants and can also be used to study more rapidly the mechanism of gene silencing. We also demonstrate that expression of two reporter genes is selectively suppressed by specific siRNA in tobacco protoplasts. In addition, we report that siRNA targeted to the AC1 gene of ACMV-[CM] specifically interfered with ACMV DNA accumulation in protoplasts and dramatically reduced the accumulation of the corresponding mRNA. Furthermore, the detection of siRNAs corresponding to the nontargeted region of the GFP gene clearly shows that siRNA-induced RNA silencing progresses along the targeted gene outside the sequence of the siRNA (also called “transitive RNA silencing”) as shown in the case of C. elegans (38) and plants (39). Thus, because basic features of PTGS also occur in protoplasts, the use of siRNAs in a protoplast system has the potential to be used for basic studies of the RNA-silencing mechanism and to identify suppression of plant virus replication to develop efficient nonspecific strategies to control viruses including ssDNA viruses.
Materials and Methods
Constructs. Construction of infectious clones of DNA-A and DNA-B of ACMV from Cameroon (ACMV-[CM]) (Fig. 1A) and East African cassava mosaic Cameroon virus (EACMCV) used in this study have been described (40, 41). Enhanced GFP (EGFP) and red fluorescent protein from Discosoma (DsRed) used in this study were from EGFP-C2 and DsRed2, respectively (CLONTECH). The plasmids pEGFP-C2 and DsRed2 were first transformed into a methylation minus Escherichia coli strain JM110. Later, the EGFP- and DsRed-coding sequences were excised as NheI and BclI fragments, ligated (Roche Diagnostics) downstream of the 35S cauliflower mosaic virus (CaMV) promoter at the AvrII and BamHI sites upstream of the nopaline synthase terminator in a pUC18-based vector, and transformed into TOP10 E. coli (Invitrogen).
Fig. 1.
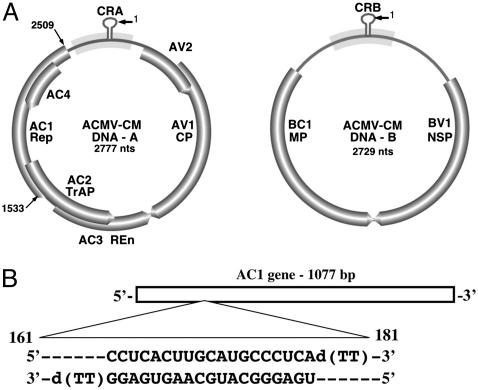
(A) Genome organization of DNA-A and DNA-B of ACMV-[CM]. DNA-A contains six ORFs: AC1–AC4, AV1, and AV2. DNA-B contains the ORFs BV1 and BC1. V represents virion sense genes, and C represents complementary sense genes. AC1 encodes replication-associated protein (Rep). (B) Schematic representation of the AC1-coding sequence of ACMV DNA-A. The siAC1 sequence (nucleotides 161–181) is as indicated. CRA, common region in DNA-A; CRB, common region in DNA-B; CP, coat protein; TrAP, transcriptional activator protein; REn, replication enhancer protein; MP, movement protein; NSP, nuclear-shuttle protein.
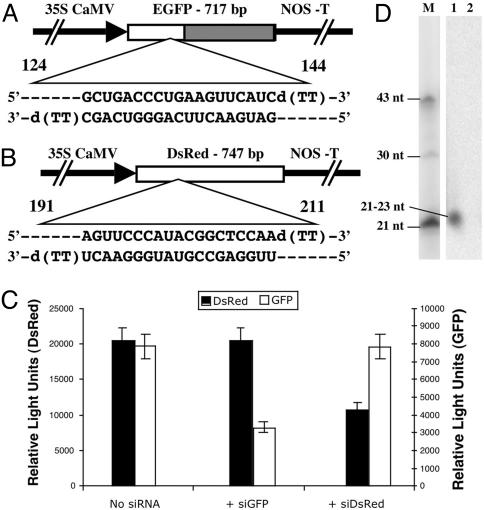
siRNA Preparation and Protoplast Transfection. siRNA corresponding to the coding regions of EGFP and DsRed2 and to the AC1 gene of ACMV were designed with 5′ phosphate, 3′ hydroxyl, and two base overhangs at the 3′ end of each strand as described (33) and chemically synthesized by Xeragon (Madison, WI). The following sequences were used: to specifically target GFP expression, siRNA cognate to GFP (siGFP) sense strand 5′-GCUGACCCUGAAGUUCAUCd(TT)-3′ (nucleotides 124–144) and antisense strand 5′-GAUGAACUUCAGGGUCAGC-d(TT)-3′ (Fig. 2A); to target DsRed2 expression, siRNA cognate to red fluorescent protein sense strand 5′-AGUUCCAGUACGGCUCCAAd(TT)-3′ (nucleotides 191–211) and antisense strand 5′-UUGGAGCCGUACUGGAACUd(TT)-3′ (Fig. 2B); and to target ACMV AC1-coding region, siRNA cognate to AC1 of ACMV-[CM] (siAC1) sense strand 5′-CCUCACUUGCAUGCCCUCAd(TT)-3′ (nucleotides 161–181) and antisense 5′-UGAGGGCAUGCAAGUGAGGd(TT)-3′ (Fig. 1B). Complementary strands were annealed by incubating at 90°C for 1 min and at room temperature for 1 h. siRNA designed to target the coding sequence of DsRed2 was kindly provided by A. Krichevsky (Harvard Medical School, Boston).
Fig. 2.
Effect of siRNA on EGFP and DsRed2 expression in BY2 tobacco protoplasts. Both the reporter plasmids were driven by the CaMV35S promoter. (A) Schematic representation of the EGFP-coding region and the siRNA sequence (nucleotides 124–144) designed to target EGFP. The slashed area represents the sequence used for the probe in D. NOS-T, nopaline synthase terminator. (B) Schematic representation of DsRed2-coding region and the corresponding siRNA sequence (nucleotides 191–211) to target DsRed2. (C) Protoplasts were transfected either with the EGFP and DsRed2 plasmid DNAs together or in combination with either siGFP or siRNA cognate to red fluorescent protein. Triplicate electroporation experiments were performed in every case. Columns and bars indicate mean and SD values, respectively. GFP and DsRed fluorescence are normalized with nontransfected control cells. (D) RNA gel blot showing siRNA accumulation in protoplasts. Lane 1, protoplasts transfected with pCaMV35S-GFP plasmid DNA with siGFP; lane 2, protoplasts transfected with pCaMV35S-GFP plasmid DNA; lane M, 21-, 30-, and 43-nt end-labeled oligos are shown as molecular size markers.
Protoplasts were isolated from 3-day-old tobacco BY2 suspension cells derived from Nicotiana tabacum L., cv. bright yellow 2 (42) and were used for transfection with viral DNA or EGFP plasmid DNA and DsRed2 plasmid DNA together with or without its cognate siRNA (43, 44). Protoplasts (1.5 million) were electroporated by using an electroporator (Bio-Rad) at 300 V, 125 μF (31) either with 5 μg each of EGFP and DsRed2 plasmid DNAs together or 4 μg each of DNA-A and DNA-B components of ACMV or EACMCV along with 20 μg of sheared herring sperm DNA with 3 μg of appropriate siRNA. After transfection, protoplasts were maintained at 28°C and harvested at different time points for DNA, RNA, and microscopic analysis.
Southern Blot and Northern Blot Analysis. Southern blotting was performed as described (45). Five micrograms of total DNA isolated from protoplasts (46) was separated by electrophoresis in a 1% agarose gel in 1× TBE (90 mM Tris-borate/2 mM EDTA, pH 8.3) without ethidium bromide and transferred to Hybond N+ membrane (Amersham Pharmacia International). For Northern blotting, 10 μg of total RNA isolated from protoplasts with an RNA isolation kit (Qiagen) was run on 1% formaldehyde agarose gels and transferred to Hybond N+ membrane. For making an ACMV-specific probe, a 794-bp EcoRI fragment (nucleotides 1789–2583) of ACMV-Ug DNA-A was used, and, for EACMCV, a 944-bp EcoRI fragment (nucleotides 1821–2765) of EACMV-Ug2 DNA-A (47) was used. These DNA fragments were labeled by using [α-32P]dCTP and a random primer-labeling kit (Prime II kit, Stratagene). Hybridization was carried out at 65°C for Southern blots and at 42°C for Northern blots. Posthybridization washes were done sequentially with 2× standard saline citrate (SSC) (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), 0.5× SSC, and 0.2× SSC along with 0.1% SDS, each for 30 min at 65°C. Blots were scanned by using a PhosphorImager and quantified by using IQMACV1.2 software (Storm, Amersham Pharmacia).
Low molecular weight RNA extraction and RNA gel blot analysis were carried out as described (8). For probe, a 0.5-kb DNA fragment between the BssSI (nucleotide 181) and BclI (nucleotide 717) of EGFP-C2 was gel-purified and labeled by using [α-32P]dCTP and a random primer-labeling kit (Prime II kit, Stratagene).
Transient-Expression Assay and Microscopic Analysis. GFP and DsRed fluorescence in protoplasts was measured by using a fluorometer (FLUOstar OPTIMA, BMG Labtechnologies, Durham, NC). Protoplasts were transfected with p35S-EGFP and p35S-DsRed plasmid DNAs together along with or without either siGFP or siRNA cognate to red fluorescent protein. Twenty-four hours posttransfection, protoplasts were collected by centrifugation at 1,000 rpm (Eppendorf 5417R) for 10 min at 28°C. Cells were resuspended in 150 μl of extraction buffer (50 mM NaHPO4 buffer, pH 7.0/10 mM EDTA/0.1% Triton X-100/0.1% sodium lauryl sarcosine/1 mM DTT) and sonicated for a total period of 14 sec with 2-sec pulses at 0.5-sec intervals. Samples were kept on ice except during the sonication and centrifuged at 12,000 rpm for 10 min at 4°C. The supernatant was transferred to a new tube and left on ice. To 50 μl of extraction buffer in each test well of a 96-well plate 40 μl of cell extract was added, mixed well, and incubated in the dark at room temperature for 30 min. GFP and DsRed fluorescence were measured after an excitation at 485 nm and emission at 520 nm for GFP and 544-nm and 580-nm excitation and emission, respectively, for DsRed.
Fluorescence microscopy with laser scanning was used to capture images. Protoplasts were inoculated with p35S-EGFP plasmid DNA with or without siGFP. The effect of siRNA on GFP expression was monitored 24 h posttransfection. To do this, protoplasts were harvested by centrifugation at 1,000 rpm for 10 min and resuspended in 500 μl of culture medium. Thirty microliters were transferred onto a glass slide and observed via microscopy. Images were recorded with equal exposure time under nonsaturated conditions for 141 randomly chosen GFP-expressing protoplasts. Gene expression was quantified in both control and targeted cells (33, 48). Quantification of pixel intensity was performed by using IMAGEJ software and normalized to a background.
Results
siRNA Interference with Reporter Gene Expression. To assess the effectiveness of siRNA in a plant cell system, we used protoplasts derived from tobacco BY2 cells and genes encoding the reporter proteins EGFP and DsRed2. The 21-nt sense and antisense ssRNAs targeted to the EGFP-C2 and DsRed2 were selected from the coding regions and designed as described in Materials and Methods. The EGFP- and DsRed2-coding sequences are 19% homologous in amino acid sequence and 36% in nucleic acid sequence. The siRNA for EGFP is 50% identical to the analogous sequence in DsRed2, whereas the siRNA for DsRed2 is 33% identical to the analogous sequence in EGFP. Each strand was synthesized separately, and pairs were annealed to create the duplex dsRNA with the characteristics of siRNA. In the initial studies we tested the effect of siRNA concentration on GFP expression. Protoplasts were transfected with the reporter GFP plasmid with or without siGFP. Transient expression of GFP in transformed protoplasts was measured by using a fluorometer at different times after transfection, and it was determined that GFP expression was maximum for 24–36 h posttransfection. We found that higher concentrations of siRNA (3 μg/ml) interfere more effectively than lower concentrations (0.5 μg/ml). Therefore, we chose 3 μg of siRNA per reaction for all subsequent experiments. Protoplasts coelectroporated with plasmids encoding GFP and DsRed plus siRNA cognate to GFP showed a significant reduction of GFP fluorescence to 58% of the level in the absence of siRNA (Fig. 2C). In contrast, siGFP did not interfere with DsRed fluorescence, indicating that interference is sequence-specific (Fig. 2C). Similarly, protoplasts cotransfected with both reporter plasmids along with siRNA targeted to DsRed showed a 47% reduction in DsRed expression (compared with control) but had no effect on GFP expression (Fig. 2C). This study demonstrated that synthetic siRNAs function in isolated plant cells and specifically interfered with the gene expression.
GFP expression in protoplasts was also monitored by using fluorescence microscopy. To quantify the effect of siRNAs on GFP expression, images of protoplasts expressing GFP were recorded with equal exposure time under nonsaturating conditions with further monitoring of green fluorescence. For comparison, at least 141 randomly chosen transfected control and targeted cells were quantified. The results revealed that GFP expression was higher in control cells compared with targeted cells, indicating that the results did not reflect reduced efficiency of transfection but were caused by interference with GFP expression.
siRNA-Induced Transitive RNA Silencing. Next we designed studies to determine whether introduced siRNA could induce RNA silencing via a transitive mechanism. Protoplasts were cotransfected with pCaMV35S-GFP plasmid DNA and siGFP targeted to the 5′ region of the GFP gene sequence. Low molecular weight RNA was isolated 24 h after electroporation. RNA gel blot analysis with specific probe designed to hybridize with the nontargeted region of GFP revealed the accumulation of siRNAs (Fig. 2D). This result indicates de novo synthesis of siRNAs in protoplasts and demonstrates the spreading along the targeted region as proposed for RNA silencing. Such molecules were not detected in protoplasts transfected with GFP plasmid DNA alone.
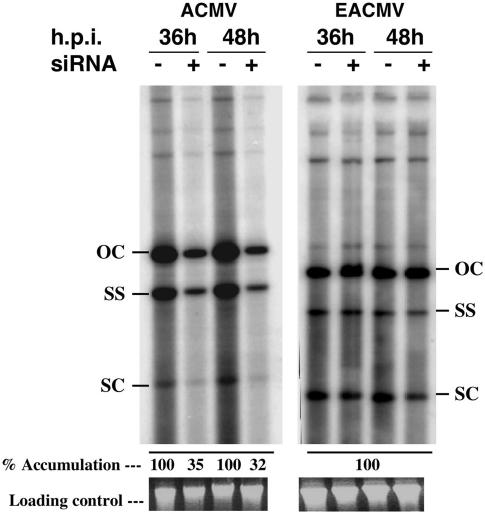
siRNA Interference on mRNA and Viral DNA Accumulation. We tested whether siRNAs are capable of reducing geminiviral DNA accumulation in protoplasts. siRNA was designed to target the AC1 gene (siAC1) encoding the replication-associated protein of ACMV-[CM]. Protoplasts were coinoculated with the combination of (i) infectious clones of ACMV DNA-A and DNA-B (together) with or without siAC1 or (ii) infectious clones of DNA-A and DNA-B of EACMCV with or without siAC1. Total DNA was isolated from protoplasts 36 and 48 h posttransfection, and viral DNA accumulation was quantified by using Southern blot hybridization. We observed a 65–68% reduction in viral DNA accumulation in reactions with siRNA compared with the control 36 and 48 h posttransfection, respectively (Fig. 3 Left), indicating that siRNAs inhibited ACMV-DNA accumulation. However, siAC1 did not interfere with accumulation of EACMCV DNA (Fig. 3 Right), another species of cassava-infecting geminiviruses, indicating that this siAC1 was highly specific in its action. The siAC1 sequence of ACMV shares 67% homology to its counterpart in EACMCV.
Fig. 3.
Effect of siRNA targeted to the AC1 of ACMV on ACMV and EACMCV DNA accumulation. Southern blots showing relative levels of viral DNA accumulation 36–48 h posttransfection in BY2 tobacco protoplasts. For cotransfection, either infectious DNA-A and DNA-B clones of ACMV or EACMCV with (+) or without (-) siAC1 were used. Five micrograms of total DNA isolated from protoplasts was loaded in each lane. The blots were probed with [α-32P]dCTP-labeled ACMV-specific (Left) or EACMV-specific (Right) DNA. Ethidium bromide-stained gel to show loading control. h.p.i., Hours postinoculation; OC, open circular; SS, single strand; SC, super-coiled, viral DNA forms.
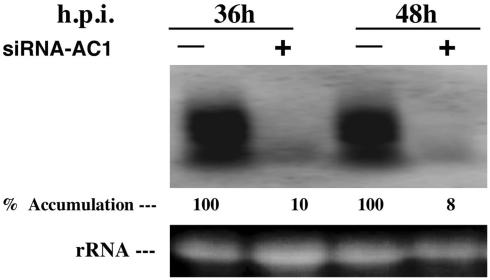
Next we examined the effect of siRNA on accumulation of viral mRNA. Total RNA isolated from protoplasts inoculated with infectious clones of DNA-A and DNA-B of ACMV with or without siAC1 was subjected to Northern blot hybridization by using AC1 gene-specific probe (Fig. 4). Surprisingly, we found a 90–92% reduction in AC1 mRNA level compared with the control 36 and 48 h posttransfection, respectively. This result indicated that siAC1 specifically degraded the polycistronic transcript, which contains the AC1 mRNA. This result demonstrates that siAC1 interfered with the stability of ACMV-specific mRNAs.
Fig. 4.
Northern blot analysis of AC1-specific mRNA in transfected BY2 tobacco protoplasts. Ten micrograms of total RNA isolated from protoplasts inoculated with the infectious clones of DNA-A and DNA-B of ACMV-[CM] with (+) or without (-) siRNA specific for AC1 of ACMV-[CM] was loaded in each lane. AC1-specific [α-32P]dCTP-labeled DNA was used as the probe. Ethidium bromide staining of ribosomal RNA (rRNA) shows equal loading of the samples. h.p.i., Hours postinoculation.
Discussion
Viruses are known to trigger PTGS with the subsequent production of virus-derived siRNAs in the case of potato virus X-infected plants (8) and Cymbidium ringspot virus in protoplasts (49). This induced defense is characterized by sequence-specific resistance against virus infection (16). Earlier studies demonstrated that gene silencing can be triggered by either the presence of multiple transgene copies in transgenic plants (9) or additional gene copies, such as high or low molecular weight dsRNAs introduced through Agrobacterium or particle bombardment (8, 28–31, 39, 50, 51). The use of siRNAs, an intermediate in the gene-silencing pathway, has become a powerful tool for specifically down-regulating gene expression, as has been demonstrated successfully in a wide variety of mammalian cells (4, 32–35) but not in plants, with the exception of a transient phenotype on GFP plants bombarded with siGFP (39). Besides, there has been no direct proof that siRNAs could specifically down-regulate gene expression and/or viral replication, particularly in plant cell cultures and specifically for ssDNA viruses. In our laboratory, we observed that geminivirus-induced PTGS is associated with the production of virus-derived siRNAs, leading to recovery phenotype in N. benthamiana and in cassava (unpublished data). This led us to ask whether gene silencing can be used as a tool to control geminivirus infection.
In this study we developed and used a protoplast system to facilitate rapid and quantitative analysis of synthetic double-stranded siRNA-mediated interference on gene expression. By electroporation of N. tabacum BY2 protoplasts, we demonstrated that siRNAs can be delivered into protoplasts, making it possible to evaluate the effect of siRNAs targeted against GFP and DsRed plasmids, and the AC1 gene of a geminivirus, ACMV-[CM]. In both cases, presence of the siRNA in the plant cells resulted in a sequence-specific down-regulation of the target gene. Similarly, codelivery of both reporter genes in combination with either one of the siRNA specifically inhibited the respective targeted gene expression without affecting the expression of the other gene. In addition, it is clear from our results that siRNA can induce gene silencing in a “transitive manner” in cultured plant cells; i.e., the targeting of siRNAs to one sequence in a gene results in degradation of the entire or most of the mRNA to short polynucleotides outside the siRNA-targeted region. This phenomenon has been demonstrated with the production of siRNAs on the 5′ side of the targeted gene in C. elegans (38) and on both sides of the targeted gene in transgenic GFP plants (39).
The knowledge that siRNA can down-regulate GFP or DsRed expression in protoplasts raised the possibility that such molecules could be used to suppress geminivirus accumulation in plant cells. In geminiviruses, the replication-associated protein AC1 is a multifunctional protein and is indispensable for viral DNA replication. It was reported that AC1 represses its own expression at the level of transcription (26, 27). siRNA directed against the AC1 gene of the geminivirus ACMV-[CM] reduced viral DNA accumulation of ACMV-[CM] by >60% 48 h posttransfection. The effect of siAC1 on accumulation of AC1 mRNA was very dramatic and showed a 90% decrease in mRNA levels. Based on the results of siGFP-induced RNA silencing, we predicted that introduction of siAC1 would result in degradation of AC1 mRNA and/or might serve as a primer to synthesize dsRNA by using the AC1 viral mRNA as the template to amplify the PTGS signal (2, 11). Once synthesized, AC1 protein is stable, and it is therefore not surprising that the effect of PTGS is less on DNA accumulation than on accumulation of mRNA level. Moreover, once viral DNA is produced, other viral ssDNA-binding proteins, including the coat protein or nuclear shuttle protein BV1, protect it. Therefore, although 90% of mRNA was degraded, the remaining 10% is probably sufficient to enable continued accumulation of viral DNA. Use of a cell-based system such as that described here for rapid evaluation and study of siRNA and the ability to down-regulate gene expression will be a valuable tool for investigating gene regulation in plants. In addition, we provide evidence that siRNA can interfere with and suppress accumulation of the economically important ssDNA geminiviruses.
As in the case of the GFP and DsRed reporter genes, siRNA-mediated gene silencing was specific against the parent sequence only, with the siAC1 proving ineffective for suppressing accumulation of EACMCV. The nucleotide sequence of the siAC1 targeted in the AC1 gene of ACMV-[CM] differs by 33% compared with its counterpart in EACMCV. ACMV siAC1 differs from the EACMCV-AC1 counterpart by 7 nt, revealing the specificity of siRNAs. We concluded that the inhibitory effect requires a very high degree of identity between the siRNAs and the target mRNA sequences.
As a counterdefense mechanism, certain plant viruses encode suppressor proteins that can block one or more steps in the RNA-silencing pathway used by plants to suppress viral accumulation (2, 18). For geminiviruses, AC2 of ACMV from Kenya (19) and C2 of TYLCCNV (23) have been reported to suppress gene silencing in plants. Furthermore, a host protein that suppresses PTGS has been identified (52). Although a number of plant RNA viral suppressors have been identified, their exact mechanism(s) of action on the RNA-silencing pathway is yet to be determined.
The use of siRNAs in protoplasts to specifically down-regulate gene expression provides a rapid way to study gene regulation and has certain advantages over plant viral vectors, some of which act as inducers and suppressors of PTGS. Our results suggest that the use of siRNA in protoplast systems may be a valuable tool for understanding the mechanism of gene silencing in plants as well as a means of determining the point in the silencing pathway at which plant viral suppressors act.
Acknowledgments
We thank Dr. Anna Krichevskey (Harvard Medical School, Boston) for kindly providing siRNA for GFP and dsRed for our initial experiments; Drs. E. Nielsen, M. Fujiki, J. C. Koo, S. Dai, and C. Menne (Donald Danforth Plant Science Center) for assistance with fluorescence experiments; and Dr. Howard Berg and Heather Ford for help in microscopy. This work was funded by the Donald Danforth Plant Science Center.
Abbreviations: PTGS, posttranscriptional gene silencing; dsRNA, double-stranded RNA; siRNA, short interfering RNA; AC1, replication-associated protein; ACMV, African cassava mosaic virus; [CM], Cameroon strain; ssDNA, single-stranded DNA; EACMCV, East African cassava mosaic Cameroon virus; EGFP, enhanced GFP; DsRed, red fluorescent protein from Discosoma; CaMV, cauliflower mosaic virus; siGFP, siRNA cognate to GFP; siAC1, siRNA cognate to AC1 of ACMV-[CM].
References
- 1.Baulcombe, D. C. (1999) Curr. Biol. 9, R599-R601. [DOI] [PubMed] [Google Scholar]
- 2.Vance, V. & Vaucheret, H. (2001) Science 292, 2277-2280. [DOI] [PubMed] [Google Scholar]
- 3.Napoli, C., Lemieux, C. & Jourgensen, R. (1990) Plant Cell 2, 279-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mellow, C. C. (1998) Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- 5.Cogoni, C. & Macino, G. (1997) Proc. Natl. Acad. Sci. USA 94, 10233-10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond, S. M., Caudy, A. A. & Hannon, G. J. (2001) Nat. Rev. Genet. 2, 110-119. [DOI] [PubMed] [Google Scholar]
- 7.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 8.Hamilton, A. J. & Baulcombe, D. C. (1999) Science 286, 950-952. [DOI] [PubMed] [Google Scholar]
- 9.Waterhouse, P. M., Wang, M. B. & Lough, T. (2001) Nature 411, 834-842. [DOI] [PubMed] [Google Scholar]
- 10.Dalmay, T., Hamilton, A., Rudd, S., Angell, S. & Baulcombe, D. C. (2000) Cell 101, 543-553. [DOI] [PubMed] [Google Scholar]
- 11.Ahlquist, P. (2002) Science 296, 1270-1273. [DOI] [PubMed] [Google Scholar]
- 12.Tang, G., Reinhart, B. J., Bartel, D. P. & Zamore, P. D. (2003) Genes Dev. 17, 49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuschl, T., Zamore, P. D., Lehmann, R., Bartel, D. P. & Sharp, P. A. (1999) Genes Dev. 13, 3191-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamore, P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. (2000) Cell 101, 25-33. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir, S. M., Lendeckel, W. & Tuschl, T. (2001) Genes Dev. 15, 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ratcliff, F. G., MacFarlane, S. A. & Baulcombe, D. C. (1999) Plant Cell 11, 1207-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, H., Li, W. X. & Ding, S. W. (2002) Science 296, 1319-1321. [DOI] [PubMed] [Google Scholar]
- 18.Ananthalakshmi, R., Pruss, G. J., Ge, X., Marathe, R., Mallory, A. C., Smith, T. H. & Vance, V. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voinnet, O., Pinto, Y. M. & Baulcombe, D. C. (1999) Proc. Natl. Acad. Sci. USA 96, 14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kjemtrup, S., Sampson, K. S., Peele, C. G., Nguyen, L. V., Conkling, M. A., Thompson, W. F. & Robertson, D. (1998) Plant J. 14, 91-100. [DOI] [PubMed] [Google Scholar]
- 21.Peele, C., Jordan, C. V., Muangsan, N., Tunage, M., Egelkrout, E., Eagle, P., Hanley-Bowdoin, L. & Robertion, D. (2001) Plant J. 27, 357-366. [DOI] [PubMed] [Google Scholar]
- 22.Turnage, M. A., Muangsan, N., Peele, G. C. & Robertson, D. (2002) Plant J. 30, 107-114. [DOI] [PubMed] [Google Scholar]
- 23.Wezel, R. V., Dong, X., Liu, H., Tien, P., Stanley, J. & Hong, Y. (2002) Mol. Plant–Microbe Interact. 15, 203-208. [DOI] [PubMed] [Google Scholar]
- 24.Brown, J. K. (1994) FAO Plant Prot. Bull. 42, 3-32. [Google Scholar]
- 25.Lapidot, M. & Friedmann, M. (2002) Ann. Appl. Biol. 140, 109-127. [Google Scholar]
- 26.Stenger, D. C., Revington, G. N., Stevenson, M. C. & Bisaro, D. M. (1991) Proc. Natl. Acad. Sci. USA 88, 8029-8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanley-Bowdoin, L., Settlage, S. B., Orozco, B. M., Nagar, S. & Robertion, D. (1999) Crit. Rev. Plant Sci. 18, 71-106. [PubMed] [Google Scholar]
- 28.Schweizer, P., Pokorny, J., Schulze-Lefert, P. & Dudler, R. (2000) Plant J. 24, 895-903. [DOI] [PubMed] [Google Scholar]
- 29.Tenllado, F. & Diaz-Ruiz, J. R. (2001) J. Virol. 75, 12288-12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanno, T., Naito, S. & Shimamoto, K. (2000) Plant Cell Physiol. 41, 321-326. [DOI] [PubMed] [Google Scholar]
- 31.Akashi, H., Miyagishi, M. & Taira, K. (2001) Antisense Nucleic Acid Drug Dev. 11, 359-367. [DOI] [PubMed] [Google Scholar]
- 32.Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H. & Schreiber, R. D. (1998) Annu. Rev. Biochem. 67, 227-264. [DOI] [PubMed] [Google Scholar]
- 33.Krichevskey, A. M. & Kosik, K. S. (2002) Proc. Natl. Acad. Sci. USA 99, 11926-11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gitlin, L., Karelsky, S. & Andino, R. (2002) Nature 418, 430-434. [DOI] [PubMed] [Google Scholar]
- 35.Jacque, J.-M., Triques, K. & Stevenson, M. (2002) Nature 418, 435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis, D. V., Hagstrom, J. E., Loomis, A. G., Wolf, J. A. & Herweijer, H. (2002) Nat. Genet. 32, 107-108. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, C. L., Jones, L., Baulcombe, D. C. & Maule, A. J. (2001) Plant J. 25, 417-425. [DOI] [PubMed] [Google Scholar]
- 38.Sijen, T., Fleenor, J., Simmer, F., Thijssen, K. L., Parrish, S., Timmons, L., Plasterk, R. H. & Fire, A. (2001) Cell 107, 465-476. [DOI] [PubMed] [Google Scholar]
- 39.Klahre, U., Crete, P., Leuenberger, S. A., Iglesias, V. A. & Mein, F., Jr. (2002) Proc. Natl. Acad. Sci. USA 99, 11981-11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fondong, V. N., Pita, J. S., Rey, M. E. C., de Kochko, A., Beachy, R. N. & Fauquet, C. M. (2000) J. Gen. Virol. 81, 287-297. [DOI] [PubMed] [Google Scholar]
- 41.Fauquet, C. M. & Stanley, J. (2003) Ann. Appl. Biol. 142, 165-189. [Google Scholar]
- 42.Nagata, T., Nemoto, Y. & Haszawa, S. (1992) Int. Rev. Cytol. 132, 1-30. [Google Scholar]
- 43.Chatterji, A., Beachy, R. N. & Fauquet, C. M. (2001) J. Biol. Chem. 276, 25631-25638. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe, Y., Meshi, T. & Okada, Y. (1987) FEBS Lett. 219, 65-69. [Google Scholar]
- 45.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 3rd Ed.
- 46.Mettler, I. J. (1987) Plant Mol. Biol. Rep. 5, 346-349. [Google Scholar]
- 47.Pita, J. S., Fondong, V. N., Sangare, A., Otim-Nape, G. W., Ogwal, S. & Fauquet, C. M. (2001) J. Gen. Virol. 82, 655-665. [DOI] [PubMed] [Google Scholar]
- 48.Reichel, C., Mathur, J., Eckes, P., Langenkemper, K., Koncz, C., Schell, J., Reiss, B. & Maas, C. (1996) Proc. Natl. Acad. Sci. USA 93, 5888-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szittya, G., Silhavy, D., Molnar, A., Havelda, Z., Lovas, A., Lakatos, L., Banfalvi, Z. & Burgyan, J. (2003) EMBO J. 22, 633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voinnet, O. & Baulcombe, D. C. (1997) Nature 387, 553. [DOI] [PubMed] [Google Scholar]
- 51.Pooggin, M., Shivaprasad, P. V., Veluthambi, K. & Hohn, T. (2003) Nat. Biotechnol. 21, 131-132. [DOI] [PubMed] [Google Scholar]
- 52.Ananthalakshmi, R., Marathe, R., Ge, X., Herr, J. M., Jr., Mau, C., Mallory, A., Pruss, G., Bowman, L. & Vance, V. B. (2000) Science 290, 142-144. [DOI] [PubMed] [Google Scholar]