Abstract
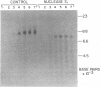
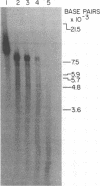
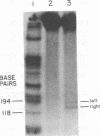
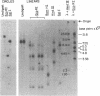
Analyses of the native DNA product of mellitin-activated avian retrovirus reverse transcription have revealed a unique structure. The vast majority of the molecules were linear, either 7.7 (genome) or 8.0 (extended genome) kilobases in length, and contained single-stranded DNA branches distributed throughout. These conclusions are based on electrophoretic properties of intact and restriction endonuclease-treated molecules before and after treatment with single-strand-specific nuclease S1. Preliminary data from linear viral DNA extracted from infected cells suggest that these molecules have a similar structure. The findings summarized in this report and those in the preceding paper indicated that the single-stranded branches are of positive polarity and are generated by a strand displacement mechanism. The existence of these branches suggests a role for strand displacement in replication and recombination.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boone L. R., Skalka A. M. Viral DNA synthesized in vitro by avian retrovirus particles permeabilized with melittin. I. Kinetics of synthesis and size of minus- and plus-strand transcripts. J Virol. 1981 Jan;37(1):109–116. doi: 10.1128/jvi.37.1.109-116.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone L. R., Skalka A. Two species of full-length cDNA are synthesized in high yield by melittin-treated avian retrovirus particles. Proc Natl Acad Sci U S A. 1980 Feb;77(2):847–851. doi: 10.1073/pnas.77.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. S., Temin H. M. Ribonucleotides in unintegrated linear spleen necrosis virus DNA. J Virol. 1980 Mar;33(3):1058–1073. doi: 10.1128/jvi.33.3.1058-1073.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett M. S., Leis J. P., Smith M. S., Faras A. J. Unwinding-like activity associated with avian retrovirus RNA-directed DNA polymerase. J Virol. 1978 May;26(2):498–509. doi: 10.1128/jvi.26.2.498-509.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina D., Benz E. W., Jr Structure of murine sarcoma virus DNA replicative intermediates synthesized in vitro. J Virol. 1980 Jan;33(1):377–389. doi: 10.1128/jvi.33.1.377-389.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni A. M., Weinberg R. A. Partially single-stranded form of free Moloney viral DNA. Nature. 1975 Jun 19;255(5510):646–648. doi: 10.1038/255646a0. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Schiff R. D. A 32,000-dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology. 1978 Aug;89(1):119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Guntaka R. V. Structure of avian tumor virus DNA intermediates. Biochem Biophys Res Commun. 1978 May 15;82(1):335–341. doi: 10.1016/0006-291x(78)90614-9. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hsu T. W., Sabran J. L., Mark G. E., Guntaka R. V., Taylor J. M. Analysis of unintegrated avian RNA tumor virus double-stranded DNA intermediates. J Virol. 1978 Dec;28(3):810–818. doi: 10.1128/jvi.28.3.810-818.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung P. P., Lee S. G. Isolation of nucleic acid-binding protein: stimulation of reverse transcriptase-catalysed DNA synthesis. Nature. 1976 Feb 12;259(5543):499–502. doi: 10.1038/259499a0. [DOI] [PubMed] [Google Scholar]
- Hunter E. The mechanism for genetic recombination in the avian retroviruses. Curr Top Microbiol Immunol. 1978;79:295–309. doi: 10.1007/978-3-642-66853-1_7. [DOI] [PubMed] [Google Scholar]
- Ju G., Boone L., Skalka A. M. Isolation and characterization of recombinant DNA clones of avian retroviruses: size heterogeneity and instability of the direct repeat. J Virol. 1980 Mar;33(3):1026–1033. doi: 10.1128/jvi.33.3.1026-1033.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis J. P., Hurwitz J. Isolation and characterization of a protein that stimulates DNA synthesis from avian myeloblastosis virus. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2331–2335. doi: 10.1073/pnas.69.8.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClements W., Hanafusa H., Tilghman S., Skalka A. Structural studies on oncornavirus-related sequences in chicken genomic DNA: two-step analyses of EcoRI and Bgl I restriction digests and tentative mapping of a ubiquitous endogenous provirus digests and tentative mapping of a ubiquitous endogenous provirus. Proc Natl Acad Sci U S A. 1979 May;76(5):2165–2169. doi: 10.1073/pnas.76.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J. C., Dobkin C., Spiegelman S. RNA primer used in synthesis of anticomplementary DNA by reverse transcriptase of avian myeloblastosis virus. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1316–1320. doi: 10.1073/pnas.77.3.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel K. P., Papas T. S., Chirikjian J. G. DNA endonucleases associated with the avian myeloblastosis virus DNA polymerase. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2659–2663. doi: 10.1073/pnas.76.6.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Hughes S. H., Kung H. J., Majors J. E., Quintrell N., Guntaka R. V., Bishop J. M., Varmus H. E. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978 Dec;15(4):1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Greenstein M., Skalka A. The circle mode of replication of bacteriophage lambda: the role of covalently closed templates and the formation of mixed catenated dimers. J Mol Biol. 1976 May 25;103(3):537–562. doi: 10.1016/0022-2836(76)90216-3. [DOI] [PubMed] [Google Scholar]
- Taylor J. M. DNA intermediates of avian RNA tumor viruses. Curr Top Microbiol Immunol. 1979;87:23–41. doi: 10.1007/978-3-642-67344-3_2. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Hsu T. W. Reverse transcription of avian sarcoma virus RNA into DNA might involve copying of the tRNA primer. J Virol. 1980 Jan;33(1):531–534. doi: 10.1128/jvi.33.1.531-534.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E., Heasley S., Kung H. J., Oppermann H., Smith V. C., Bishop J. M., Shank P. R. Kinetics of synthesis, structure and purification of avian sarcoma virus-specific DNA made in the cytoplasm of acutely infected cells. J Mol Biol. 1978 Mar 25;120(1):55–82. doi: 10.1016/0022-2836(78)90295-4. [DOI] [PubMed] [Google Scholar]
- Weinberg R. A. Structure of the intermediates leading to the integrated provirus. Biochim Biophys Acta. 1977 Mar 21;473(1):39–55. doi: 10.1016/0304-419x(77)90006-3. [DOI] [PubMed] [Google Scholar]
- Wiegand R. C., Godson G. N., Radding C. M. Specificity of the S1 nuclease from Aspergillus oryzae. J Biol Chem. 1975 Nov 25;250(22):8848–8855. [PubMed] [Google Scholar]