Abstract
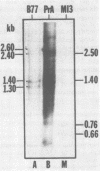
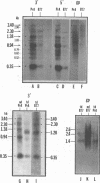
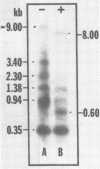
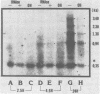
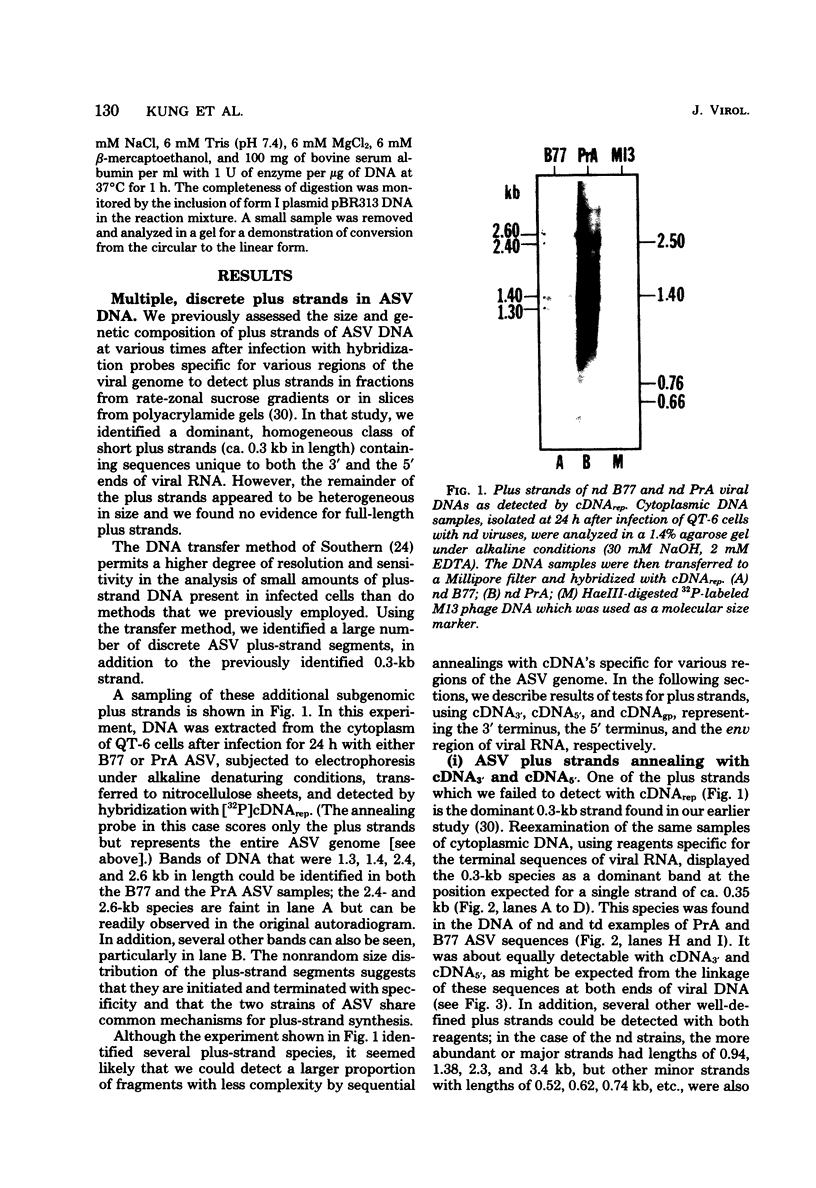
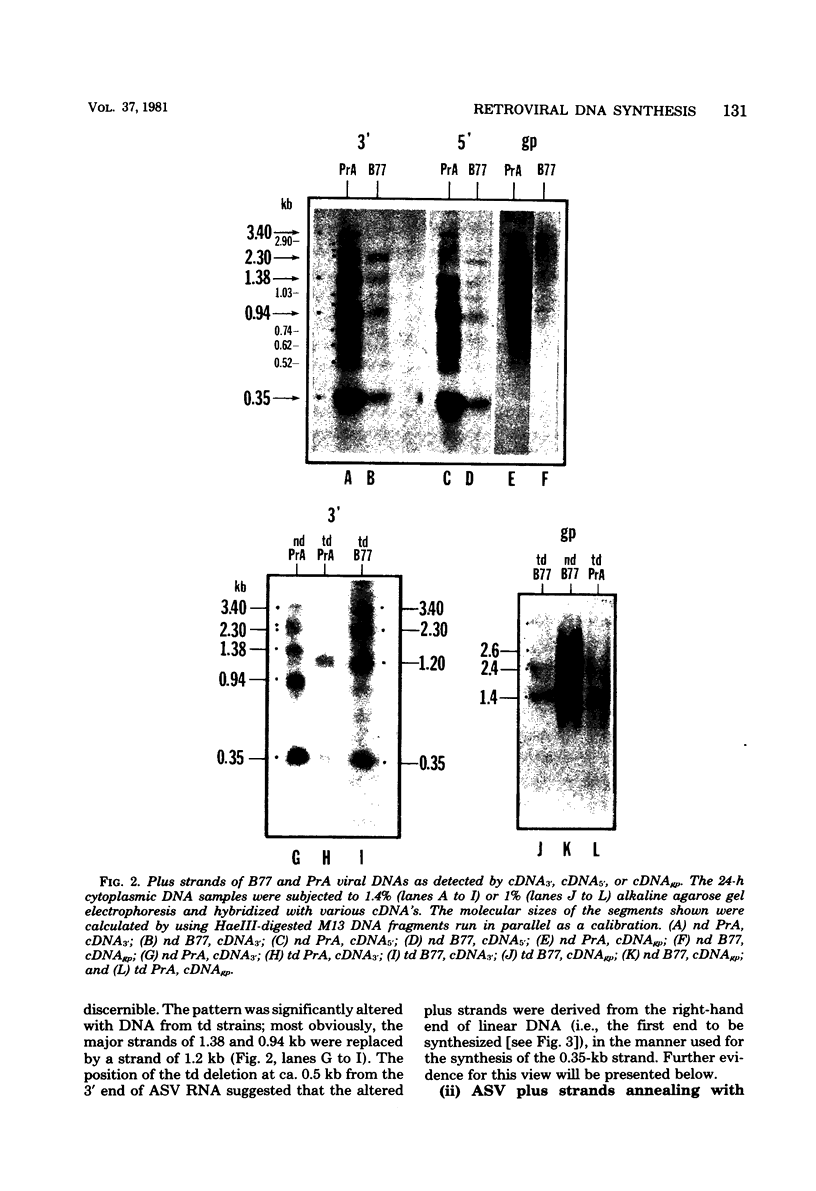
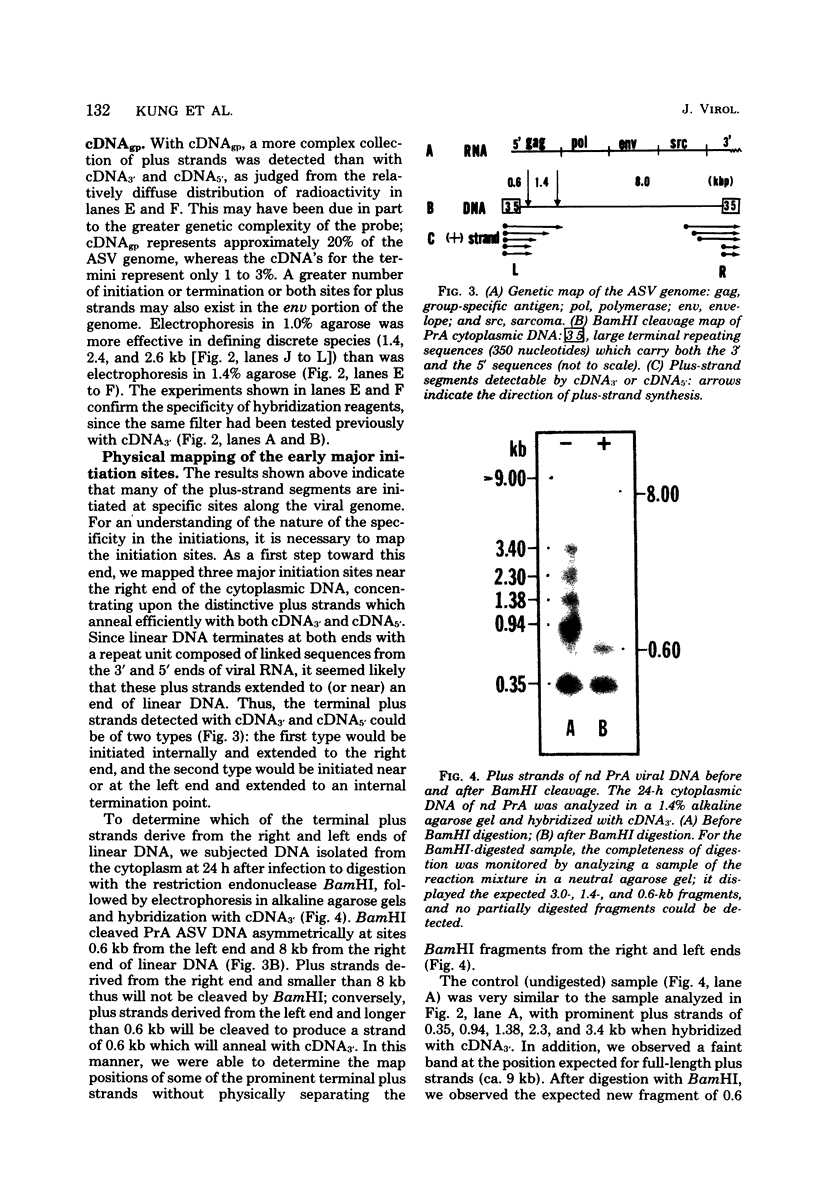
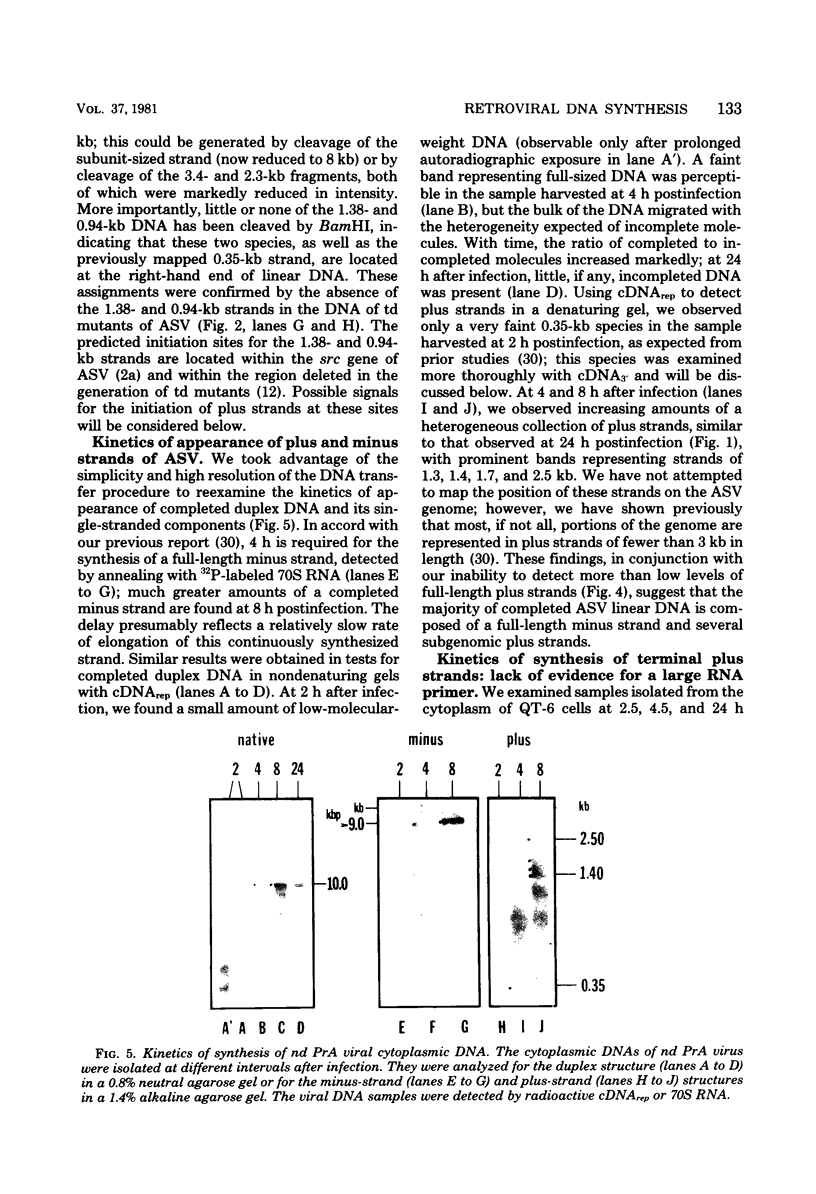
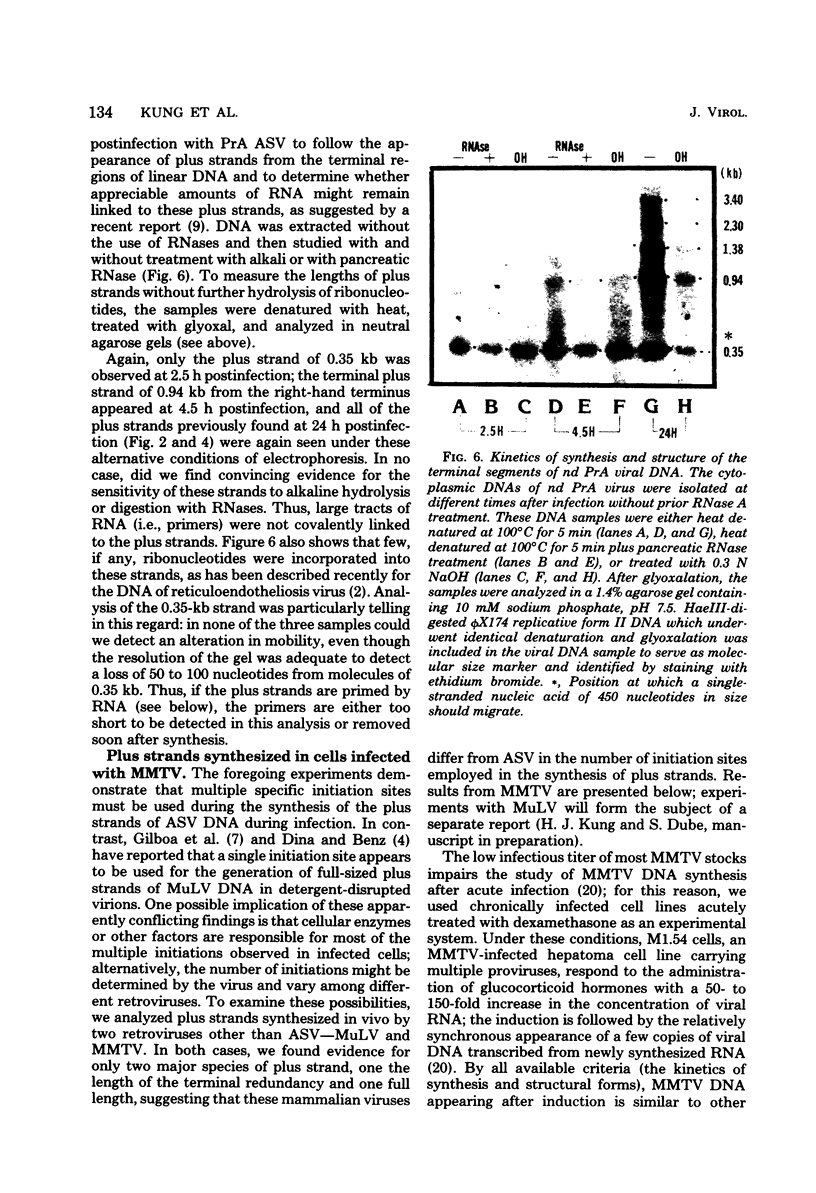
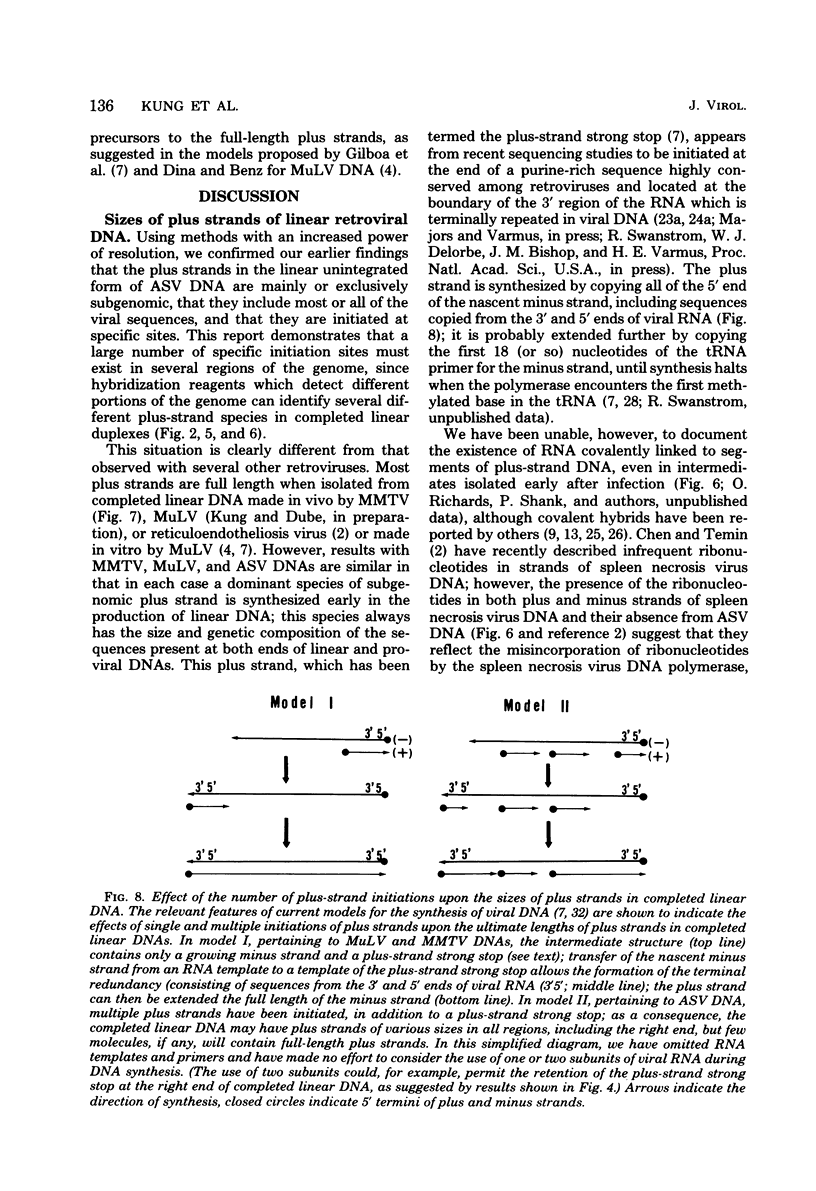
The vast majority of plus strands synthesized in quail cells acutely infected with avian sarcoma virus were subgenomic in size, generally less than 3 kilobases (kb). A series of discrete species could be identified after agarose gel electrophoresis by annealing with various complementary DNAs, indicating specificity in the initiation and termination of plus strands. The first plus strand to appear (within 2 h postinfection) was similar in length to the long redundancy at the ends of linear DNA (0.35 kb), and it annealed with complementary DNAs specific for the 3' and 5' termini of viral RNA (Varmus et al., J. Mol. Biol. 120:50-82, 1978). Several subgenomic plus-strand fragments (0.94, 1.38, 2.3, and 3.4 kb) annealed with these reagents. At least the 0.94- and 1.38-kb strands were located at the same end of linear DNA as the 0.35-kb strand, indicating that multiple specific sites for initiation were employed to generate strands which overlapped on the structural map. We were unable to detect RNA liked to plus strands isolated as early as 2.5 h postinfection; thus, the primers must be short (fewer than 50 to 100 nucleotides), rapidly removed, or not composed of RNA. To determine whether multiple priming events are a general property of retroviral DNA synthesis in vivo, we also examined plus strands of mouse mammary tumor virus DNA in chronically infected rat cells after induction of RNA and subsequent DNA synthesis with dexamethasone. In this case, multiple, discrete subgenomic DNA plus strands were not found when the same methods applied to avian sarcoma virus DNA were used; instead, the plus strands present in the linear DNA of mouse mammary tumor virus fell mainly into two classes: (i) strands of ca. 1.3 kb which appeared early in synthesis and were similar in size and genetic content to the terminally repeated sequence in linear DNA; and (ii) plus strands of the same length as linear DNA. A heterogeneous population of other strands diminished with time, was not found in completed molecules, and was probably composed of strands undergoing elongation. These two retroviruses thus appear to differ with respect to both the number of priming sites used for the synthesis of plus strands and the abundance of full-length plus strands. On the other hand the major subgenomic plus strand of mouse mammary tumor virus DNA (1.3 kb) is probably the functional homolog of a major subgenomic plus strand of avian sarcoma virus DNA (0.35 kb). The significance of this plus strand species is discussed in the context of current models which hold that it is used as a template for the completion of the minus strand, thereby generating the long terminal redundancy.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D., Gilboa E., Rothenberg E., Yoshimura F. Production of a discrete, infectious, double-stranded DNA by reverse transcription in virions of Moloney murine leukemia virus. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):869–874. doi: 10.1101/sqb.1979.043.01.093. [DOI] [PubMed] [Google Scholar]
- Chen I. S., Temin H. M. Ribonucleotides in unintegrated linear spleen necrosis virus DNA. J Virol. 1980 Mar;33(3):1058–1073. doi: 10.1128/jvi.33.3.1058-1073.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czernilofsky A. P., Levinson A. D., Varmus H. E., Bishop J. M., Tischer E., Goodman H. M. Nucleotide sequence of an avian sarcoma virus oncogene (src) and proposed amino acid sequence for gene product. Nature. 1980 Sep 18;287(5779):198–203. doi: 10.1038/287198a0. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina D., Benz E. W., Jr Structure of murine sarcoma virus DNA replicative intermediates synthesized in vitro. J Virol. 1980 Jan;33(1):377–389. doi: 10.1128/jvi.33.1.377-389.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich R., Kung H. J., Baker B., Varmus H. E., Goodman H. M., Bishop J. M. Characterization of DNA complementary to nucleotide sequences at the 5'-terminus of the avian sarcoma virus genome. Virology. 1977 Jun 1;79(1):198–215. doi: 10.1016/0042-6822(77)90345-2. [DOI] [PubMed] [Google Scholar]
- Gianni A. M., Weinberg R. A. Partially single-stranded form of free Moloney viral DNA. Nature. 1975 Jun 19;255(5510):646–648. doi: 10.1038/255646a0. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Goff S., Shields A., Yoshimura F., Mitra S., Baltimore D. In vitro synthesis of a 9 kbp terminally redundant DNA carrying the infectivity of Moloney murine leukemia virus. Cell. 1979 Apr;16(4):863–874. doi: 10.1016/0092-8674(79)90101-6. [DOI] [PubMed] [Google Scholar]
- Green M., Gerard G. F. RNA-directed DNA polymerase--properties and functions in oncogenic RNA viruses and cells. Prog Nucleic Acid Res Mol Biol. 1974;14(0):187–334. [PubMed] [Google Scholar]
- Guntaka R. V. Structure of avian tumor virus DNA intermediates. Biochem Biophys Res Commun. 1978 May 15;82(1):335–341. doi: 10.1016/0006-291x(78)90614-9. [DOI] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T. W., Sabran J. L., Mark G. E., Guntaka R. V., Taylor J. M. Analysis of unintegrated avian RNA tumor virus double-stranded DNA intermediates. J Virol. 1978 Dec;28(3):810–818. doi: 10.1128/jvi.28.3.810-818.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghans R. P., Hu S., Knight C. A., Davidson N. Heteroduplex analysis of avian RNA tumor viruses. Proc Natl Acad Sci U S A. 1977 Feb;74(2):477–481. doi: 10.1073/pnas.74.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis J., Schincariol A., Ishizaki R., Hurwitz J. RNA-dependent DNA polymerase activity of RNA tumor viruses. V. Rous sarcoma virus single-stranded RNA-DNA covalent hybrids in infected chicken embryo fibroblast cells. J Virol. 1975 Mar;15(3):484–489. doi: 10.1128/jvi.15.3.484-489.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Mitra S. W., Goff S., Gilboa E., Baltimore D. Synthesis of a 600-nucleotide-long plus-strand DNA by virions of Moloney murine leukemia virus. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4355–4359. doi: 10.1073/pnas.76.9.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscovici C., Moscovici M. G., Jimenez H., Lai M. M., Hayman M. J., Vogt P. K. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell. 1977 May;11(1):95–103. doi: 10.1016/0092-8674(77)90320-8. [DOI] [PubMed] [Google Scholar]
- Myers J. C., Dobkin C., Spiegelman S. RNA primer used in synthesis of anticomplementary DNA by reverse transcriptase of avian myeloblastosis virus. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1316–1320. doi: 10.1073/pnas.77.3.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintrell N., Varmus H. E., Bishop J. M., Nicholson M. O., McAllister R. M. Homologies among the nucleotide sequences of the genomes of C-type viruses. Virology. 1974 Apr;58(2):568–575. doi: 10.1016/0042-6822(74)90090-7. [DOI] [PubMed] [Google Scholar]
- Ringold G. M., Cardiff R. D., Varmus H. E., Yamamoto K. R. Infection of cultured rat hepatoma cells by mouse mammary tumor virus. Cell. 1977 Jan;10(1):11–18. doi: 10.1016/0092-8674(77)90134-9. [DOI] [PubMed] [Google Scholar]
- Ringold G. M., Shank P. R., Yamamoto K. R. Production of unintegrated mouse mammary tumor virus DNA in infected rat hepatoma cells is a secondary action of dexamethasone. J Virol. 1978 Apr;26(1):93–101. doi: 10.1128/jvi.26.1.93-101.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Cohen J. C., Varmus H. E., Yamamoto K. R., Ringold G. M. Mapping of linear and circular forms of mouse mammary tumor virus DNA with restriction endonucleases: evidence for a large specific deletion occurring at high frequency during circularization. Proc Natl Acad Sci U S A. 1978 May;75(5):2112–2116. doi: 10.1073/pnas.75.5.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Hughes S. H., Kung H. J., Majors J. E., Quintrell N., Guntaka R. V., Bishop J. M., Varmus H. E. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978 Dec;15(4):1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Shinnick T. M., Verma I. M., Lerner R. A. Nucleotide sequence of Moloney leukemia virus: 3' end reveals details of replications, analogy to bacterial transposons, and an unexpected gene. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3302–3306. doi: 10.1073/pnas.77.6.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveda M. M., Fields B. N., Soeiro R. Host restriction of friend leukemia virus; fate of input virion RNA. Cell. 1974 Aug;2(4):271–277. doi: 10.1016/0092-8674(74)90021-x. [DOI] [PubMed] [Google Scholar]
- Takano T., Hatanaka M. Fate of viral RNA of murine leukemia virus after infection. Proc Natl Acad Sci U S A. 1975 Jan;72(1):343–347. doi: 10.1073/pnas.72.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal J., Kung H. J., Varmus H. E., Bishop J. M. Characterization of DNA complementary to nucleotide sequences adjacent to poly(A) at the 3'-terminus of the avian sarcoma virus genome. Virology. 1977 Jun 1;79(1):183–197. doi: 10.1016/0042-6822(77)90344-0. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Illmensee R. Site on the RNA of an avian sarcoma virus at which primer is bound. J Virol. 1975 Sep;16(3):553–558. doi: 10.1128/jvi.16.3.553-558.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E., Heasley S., Kung H. J., Oppermann H., Smith V. C., Bishop J. M., Shank P. R. Kinetics of synthesis, structure and purification of avian sarcoma virus-specific DNA made in the cytoplasm of acutely infected cells. J Mol Biol. 1978 Mar 25;120(1):55–82. doi: 10.1016/0022-2836(78)90295-4. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Heasley S., Linn J., Wheeler K. Use of alkaline sucrose gradients in a zonal rotor to detect integrated and unintegrated avian sarcoma virus-specific DNA in cells. J Virol. 1976 May;18(2):574–585. doi: 10.1128/jvi.18.2.574-585.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E., Shank P. R., Hughes S. E., Kung H. J., Heasley S., Majors J., Vogt P. K., Bishop J. M. Synthesis, structure, and integration of the DNA of RNA tumor viruses. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):851–864. doi: 10.1101/sqb.1979.043.01.091. [DOI] [PubMed] [Google Scholar]
- Verma I. M. Genome organization of RNA tumor viruses. I. In vitro synthesis of full-genome-length single-stranded and double-stranded viral DNA transcripts. J Virol. 1978 Jun;26(3):615–629. doi: 10.1128/jvi.26.3.615-629.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson K. F., Schendel P. L., Rosok M. J., Ramsey L. R. Model RNA-directed DNA synthesis by avian myeloblastosis virus DNA polymerase and its associated RNase H. Biochemistry. 1979 Jul 24;18(15):3210–3219. doi: 10.1021/bi00582a004. [DOI] [PubMed] [Google Scholar]
- Weinberg R. A. Structure of the intermediates leading to the integrated provirus. Biochim Biophys Acta. 1977 Mar 21;473(1):39–55. doi: 10.1016/0304-419x(77)90006-3. [DOI] [PubMed] [Google Scholar]