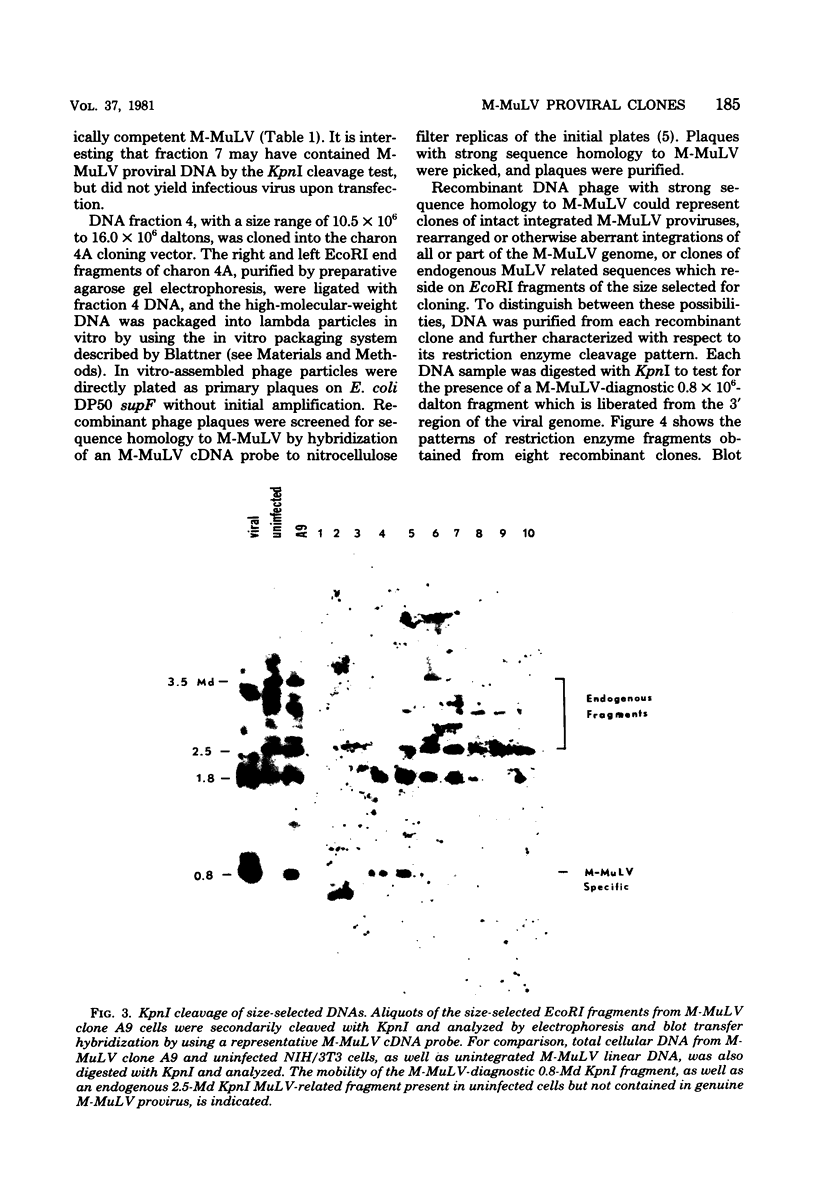
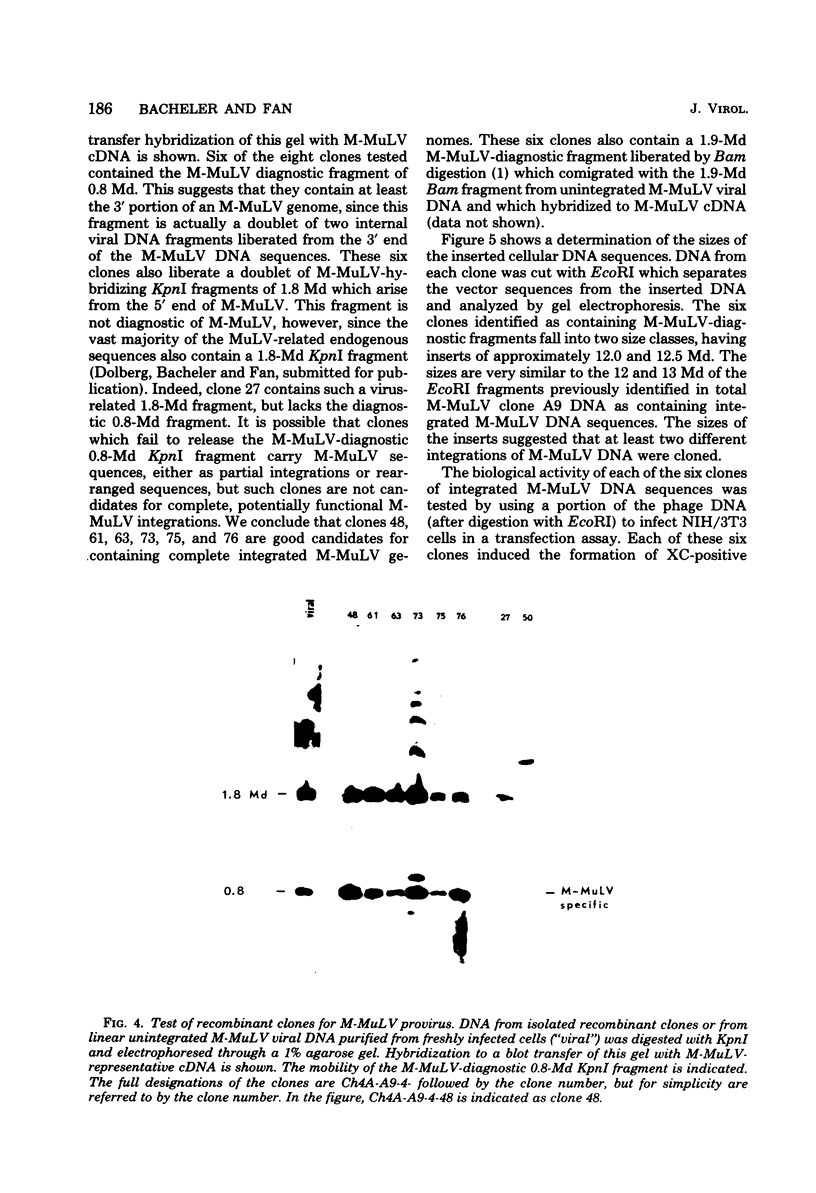
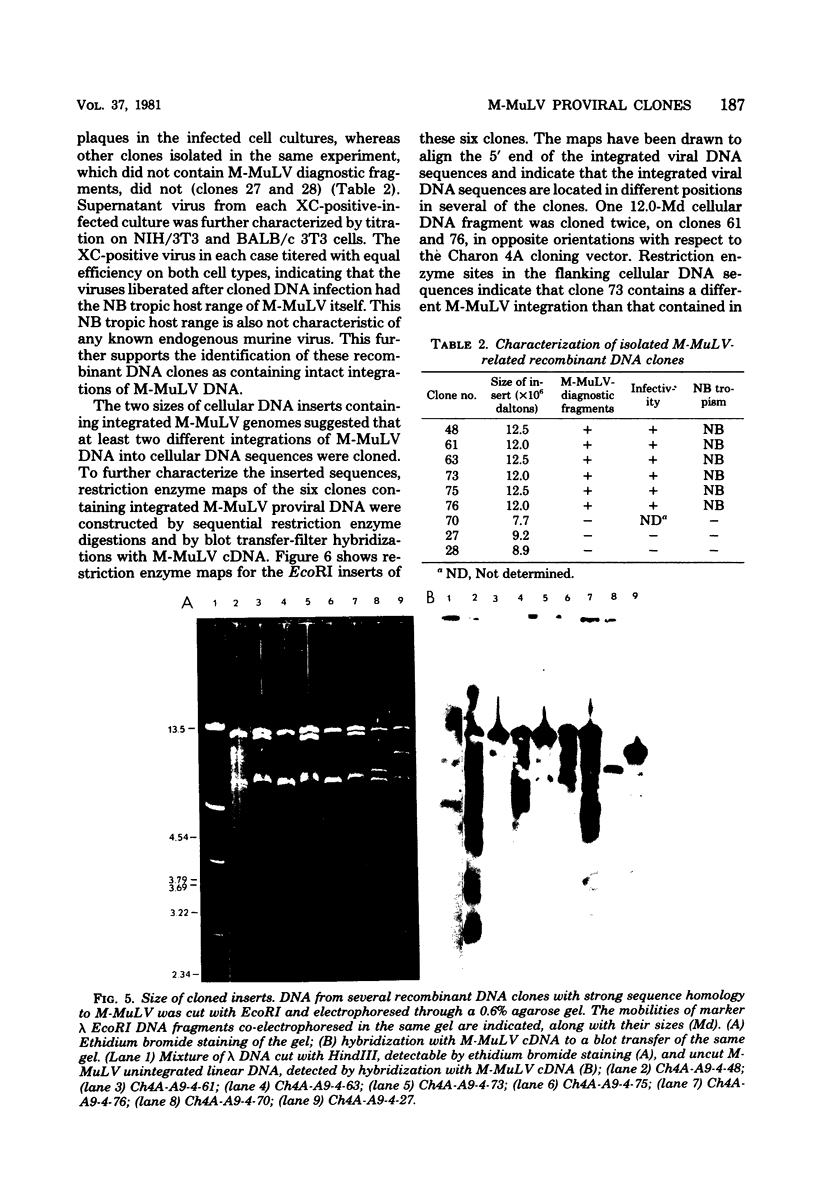
Abstract
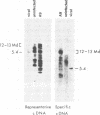
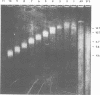
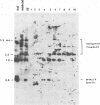
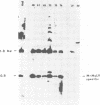
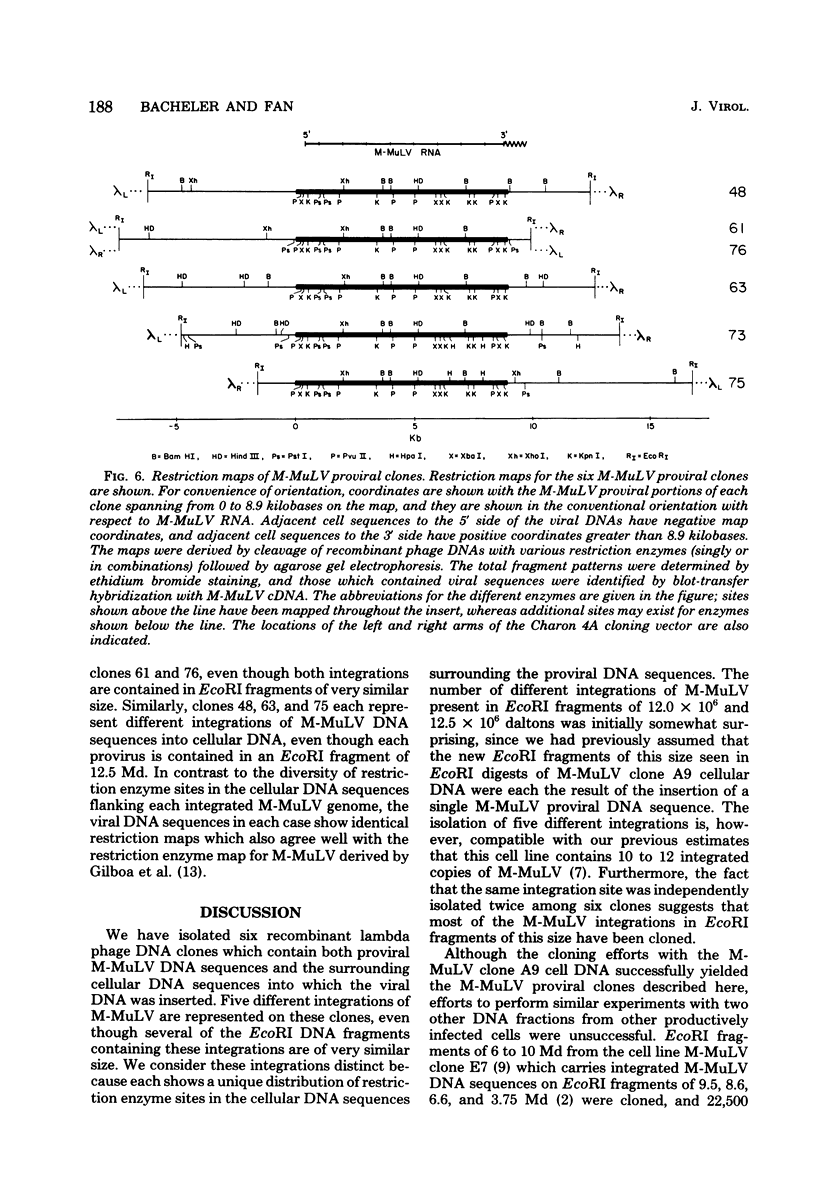
EcoRI DNA fragments from a Moloney murine leukemia virus (M-MuLV)-infected mouse fibroblast line (M-MuLV clone A9) were cloned in lambda phage Charon 4A cloning vector to derive clones containing integrated M-MuLV proviral DNA. A 10- to 16-megadalton class of EcoRI fragments was chosen for cloning, based on (i) its ability to induce XC-positive virus upon transfection of NIH/3T3 cells, and (ii) its content of a 0.8-megadalton viral KpnI fragment diagnostic for M-MuLV. Six recombinant DNA clones were isolated which contain a complete M-MuLV provirus, as judged by (i) restriction endonuclease mapping and (ii) the fact that all of the clones gave rise to XC-positive, NB-tropic virus upon DNA infection in NIH/3T3 cells. The sizes of the inserts were 12.0 (for three clones) or 12.5 megadaltons (for three clones). Restriction mapping indicated that these six clones represent five different M-MuLV proviral integrations into different cellular DNA sites.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bacheler L. T., Fan H. Integrated Moloney murine leukemia virus DNA studied by using complementary DNA which does not recognize endogenous related sequences. J Virol. 1980 Mar;33(3):1074–1082. doi: 10.1128/jvi.33.3.1074-1082.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacheler L. T., Fan H. Multiple integration sites for Moloney murine leukemia virus in productively infected mouse fibroblasts. J Virol. 1979 Jun;30(3):657–667. doi: 10.1128/jvi.30.3.657-667.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battula N., Temin H. M. Infectious DNA of spleen necrosis virus is integrated at a single site in the DNA of chronically infected chicken fibroblasts. Proc Natl Acad Sci U S A. 1977 Jan;74(1):281–285. doi: 10.1073/pnas.74.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battula N., Temin H. M. Sites of integration of infectious DNA of avian reticuloendotheliosis viruses in different avian cellular DNAs. Cell. 1978 Feb;13(2):387–398. doi: 10.1016/0092-8674(78)90207-6. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Williams B. G., Blechl A. E., Denniston-Thompson K., Faber H. E., Furlong L., Grunwald D. J., Kiefer D. O., Moore D. D., Schumm J. W. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science. 1977 Apr 8;196(4286):161–169. doi: 10.1126/science.847462. [DOI] [PubMed] [Google Scholar]
- Breindl M., Bacheler L., Fan H., Jaenisch R. Chromatin conformation of integrated Moloney leukemia virus DNA sequences in tissues of BALB/Mo mice and in virus-infected cell lines. J Virol. 1980 May;34(2):373–382. doi: 10.1128/jvi.34.2.373-382.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaani E., Aaronson S. A. Restriction enzyme analysis of mouse cellular type C viral DNA: emergence of new viral sequences in spontaneous AKR/J lymphomas. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1677–1681. doi: 10.1073/pnas.76.4.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Jaenisch R., MacIsaac P. Low-multiplicity infection of Moloney murine leukemia virus in mouse cells: effect on number of viral DNA copies and virus production in producer cells. J Virol. 1978 Dec;28(3):802–809. doi: 10.1128/jvi.28.3.802-809.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Paskind M. Measurement of the sequence complexity of cloned Moloney murine leukemia virus 60 to 70S RNA: evidence for a haploid genome. J Virol. 1974 Sep;14(3):421–429. doi: 10.1128/jvi.14.3.421-429.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Verma I. M. Size analysis and relationship of murine leukemia virus-specific mRNA's: evidence for transposition of sequences during synthesis and processing of subgenomic mRNA. J Virol. 1978 May;26(2):468–478. doi: 10.1128/jvi.26.2.468-478.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni A. M., Hutton J. R., Smotkin D., Weinberg R. A. Proviral DNA of Moloney leukemia virus: purification and visualization. Science. 1976 Feb 13;191(4227):569–571. doi: 10.1126/science.1251192. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Goff S., Shields A., Yoshimura F., Mitra S., Baltimore D. In vitro synthesis of a 9 kbp terminally redundant DNA carrying the infectivity of Moloney murine leukemia virus. Cell. 1979 Apr;16(4):863–874. doi: 10.1016/0092-8674(79)90101-6. [DOI] [PubMed] [Google Scholar]
- Gilmer T. M., Parsons J. T. Analysis of cellular integration sites in avian sarcoma virus infected duck embryo cells. J Virol. 1979 Dec;32(3):762–769. doi: 10.1128/jvi.32.3.762-769.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hughes S. H., Shank P. R., Spector D. H., Kung H. J., Bishop J. M., Varmus H. E., Vogt P. K., Breitman M. L. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978 Dec;15(4):1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- Jaenisch R. Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1260–1264. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jähner D., Jaenisch R. Integration of Moloney leukaemia virus into the germ line of mice: correlation between site of integration and virus activation. Nature. 1980 Oct 2;287(5781):456–458. doi: 10.1038/287456a0. [DOI] [PubMed] [Google Scholar]
- Keshet E., Temin H. M. Sites of integration of reticuloendotheliosis virus DNA in chicken DNA. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3372–3376. doi: 10.1073/pnas.75.7.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. H., Verma I. M. Genome organization of retroviruses. VII. Infection by double-stranded DNA synthesized in vitro from Moloney murine leukemia virus generates a virus indistinguishable from the original virus used in reverse transcription. Virology. 1980 Jan 15;100(1):194–198. doi: 10.1016/0042-6822(80)90567-x. [DOI] [PubMed] [Google Scholar]
- Lowy D. R., Rands E., Chattopadhyay S. K., Garon C. F., Hager G. L. Molecular cloning of infectious integrated murine leukemia virus DNA from infected mouse cells. Proc Natl Acad Sci U S A. 1980 Jan;77(1):614–618. doi: 10.1073/pnas.77.1.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rear J. J., Mizutani S., Hoffman G., Fiandt M., Temin H. M. Infectious and noninfectious recombinant clones of the provirus of SNV differ in cellular DNA and are apparently the same in viral DNA. Cell. 1980 Jun;20(2):423–430. doi: 10.1016/0092-8674(80)90628-5. [DOI] [PubMed] [Google Scholar]
- Rowe W. P., Pugh W. E., Hartley J. W. Plaque assay techniques for murine leukemia viruses. Virology. 1970 Dec;42(4):1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steffen D., Weinberg R. A. The integrated genome of murine leukemia virus. Cell. 1978 Nov;15(3):1003–1010. doi: 10.1016/0092-8674(78)90284-2. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Tiemeier D., Enquist L. In vitro packaging of a lambda Dam vector containing EcoRI DNA fragments of Escherichia coli and phage P1. Gene. 1977 May;1(3-4):255–280. doi: 10.1016/0378-1119(77)90049-x. [DOI] [PubMed] [Google Scholar]
- Vande Woude G. F., Oskarsson M., Enquist L. W., Nomura S., Sullivan M., Fischinger P. J. Cloning of integrated Moloney sarcoma proviral DNA sequences in bacteriophage lambda. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4464–4468. doi: 10.1073/pnas.76.9.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]