Abstract
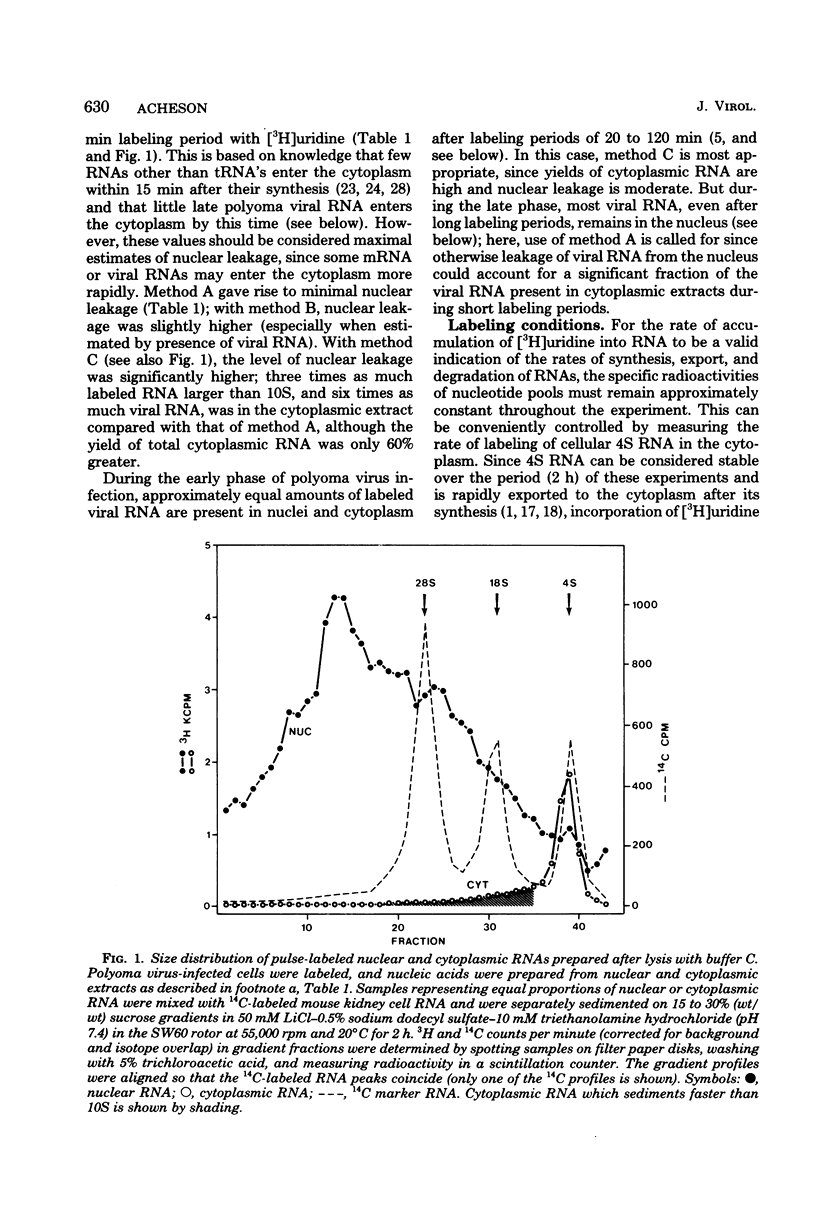
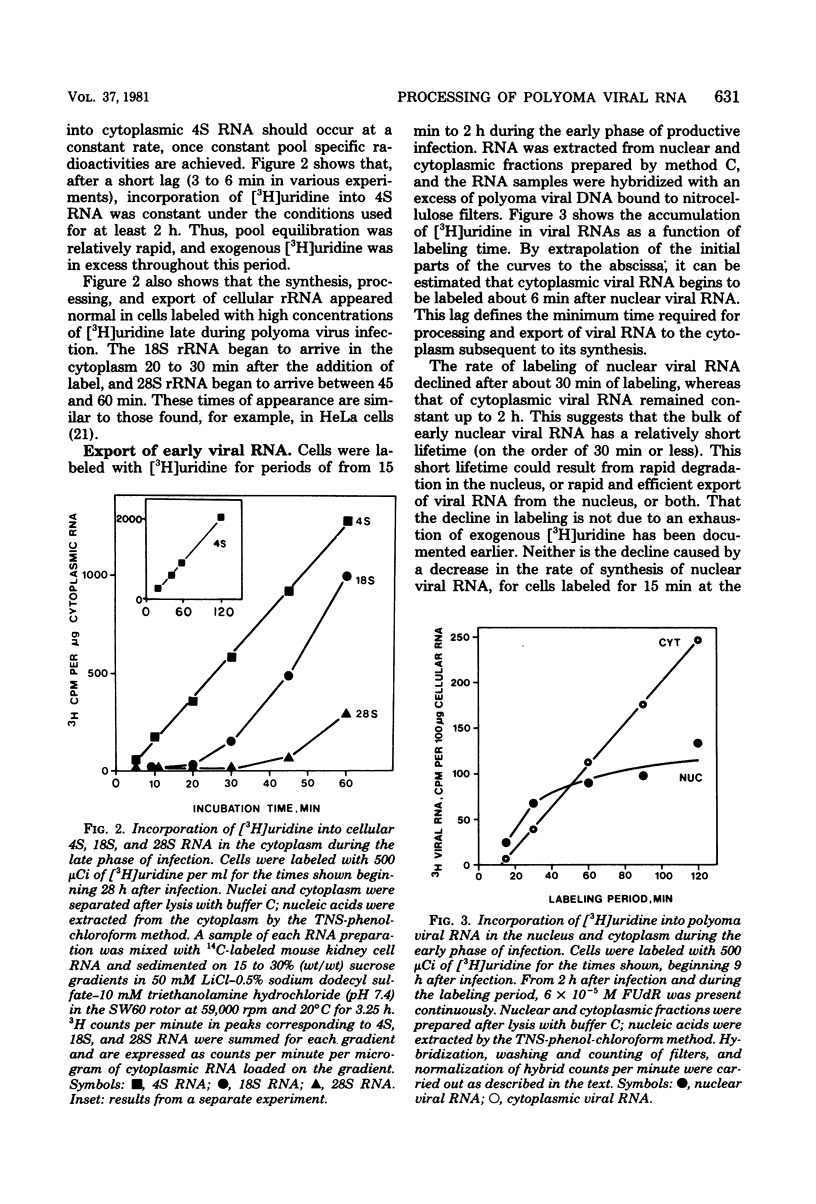
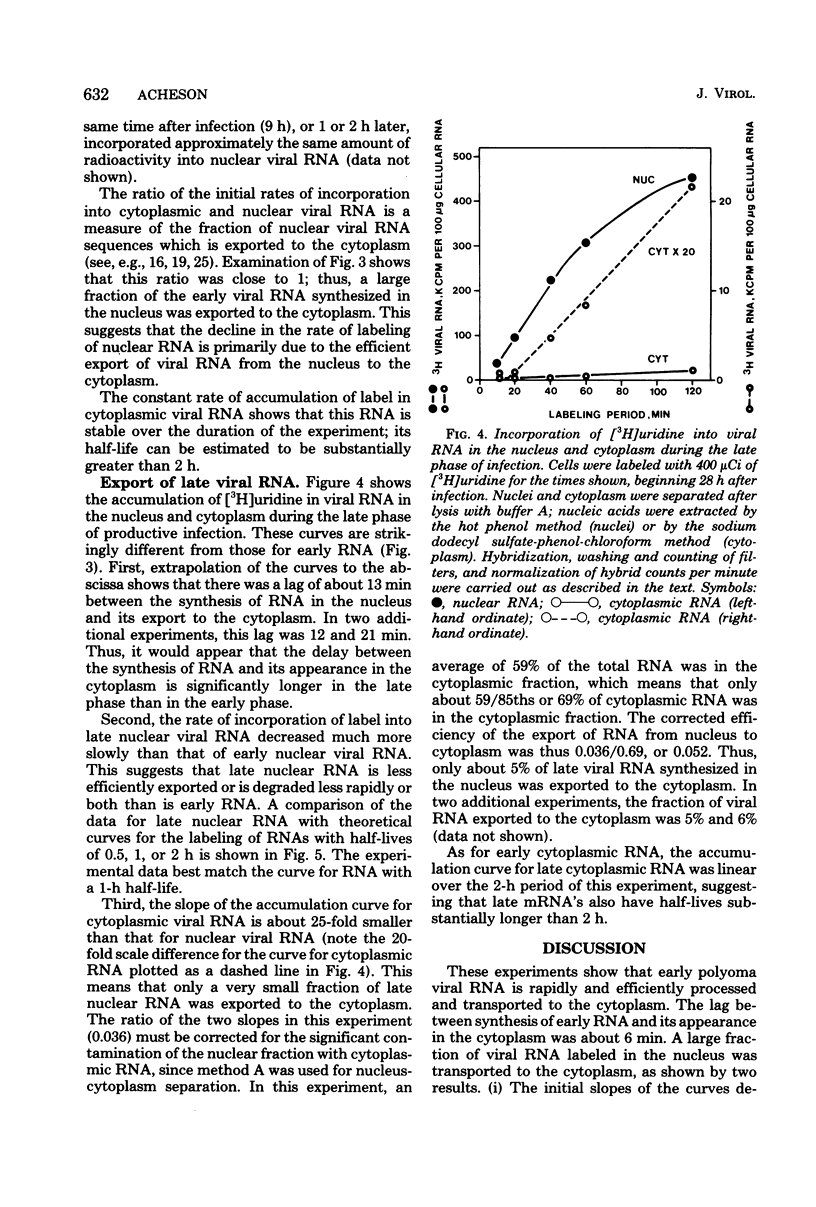
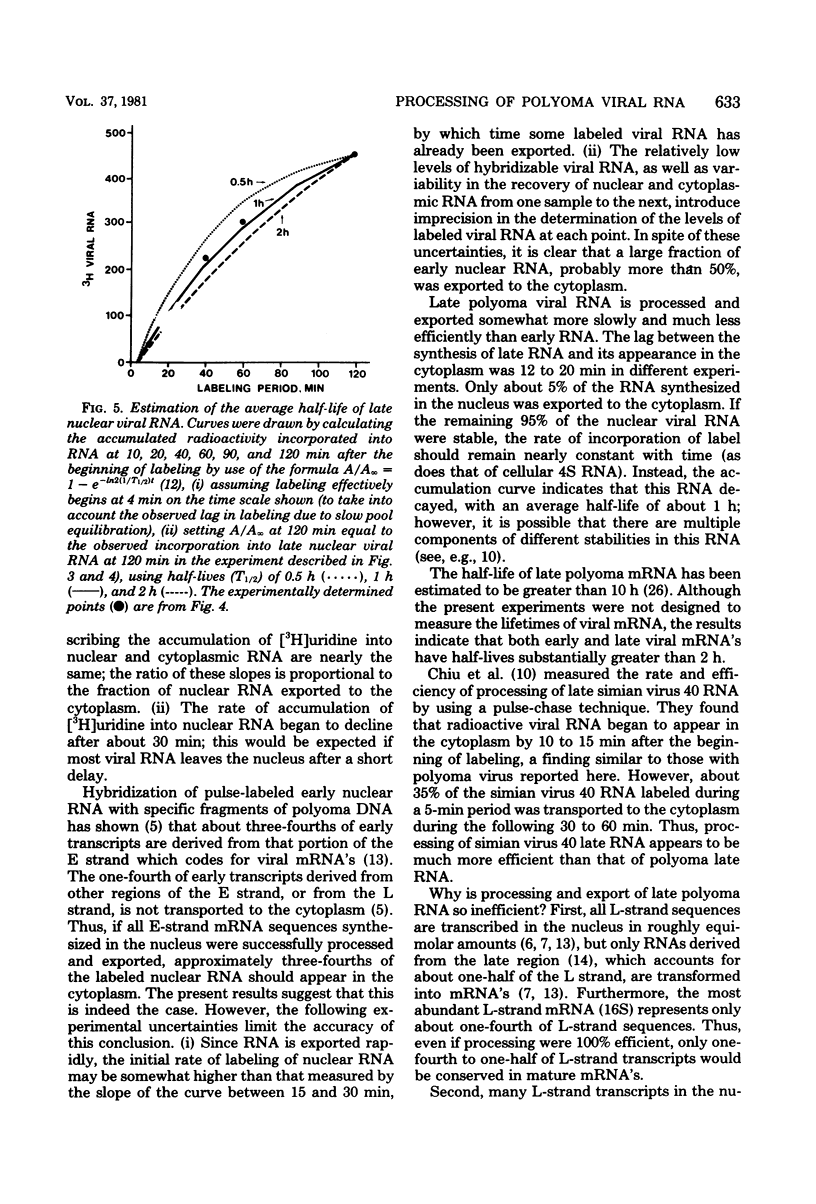
The efficiency of processing of polyoma viral RNA and of its export from nucleus to cytoplasm was measured in primary mouse kidney cells by comparing the initial rates of incorporation of [3H]uridine into cytoplasmic and nuclear viral RNA. Appropriate methods of cell fractionation were chosen to maximize yields of cytoplasmic RNA and to minimize leakage of nuclear RNA. Incorporation of [3H]uridine into cellular 4S RNA in the cytoplasm was followed to monitor pool equilibration and maintenance of an excess of radioactive precursor throughout the experimental period. During the early phase of infection (9 to 11 h, in the presence of 5-fluorodeoxyuridine), viral RNA was rapidly and efficiently exported from nucleus to cytoplasm. Viral RNA appeared in the cytoplasm within 6 min of its synthesis, greater than half of the viral RNA synthesized in the nucleus was exported to the cytoplasm. In contrast, during the late phase of infection (28 to 30 h), viral RNA was exported more slowly, appearing in the cytoplasm 12 to 20 min after its synthesis, and much less efficiently-only 5% of late nuclear transcripts was exported. The poor efficiency of processing of late viral RNA may be, in part, a result of (i) the presence in nuclear transcripts of non-mRNA sequences which are removed during processing; (ii) the presence in nuclear transcripts of multiple copies of mRNA sequences, only one of which is incorporated into mature mRNA; and (iii) inefficient polyadenylation of viral nuclear RNA.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Acheson N. H., Buetti E., Scherrer K., Weil R. Transcription of the polyoma virus genome: synthesis and cleavage of giant late polyoma-specific RNA. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2231–2235. doi: 10.1073/pnas.68.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson N. H., Miéville F. Extent of transcription of the E strand of polyoma virus DNA during the early phase of productive infection. J Virol. 1978 Dec;28(3):885–894. doi: 10.1128/jvi.28.3.885-894.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson N. H. Polyoma virus giant RNAs contain tandem repeats of the nucleotide sequence of the entire viral genome. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4754–4758. doi: 10.1073/pnas.75.10.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni Y., Locker H. Symmetrical in vivo transcription of polyoma DNA and the separation of self-complementary viral and cell RNA. Virology. 1973 Aug;54(2):495–505. doi: 10.1016/0042-6822(73)90159-1. [DOI] [PubMed] [Google Scholar]
- Beard P., Acheson N. H., Maxwell I. H. Strand-specific transcription of polyoma virus DNA-early in productive infection and in transformed cells. J Virol. 1975 Jan;17(1):20–26. doi: 10.1128/jvi.17.1.20-26.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birg F., Favaloro J., Kamen R. Analysis of polyoma virus nuclear RNA by mini-blot hybridization. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3138–3142. doi: 10.1073/pnas.74.8.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buetti E. Characterization of late polyoma mRNA. J Virol. 1974 Aug;14(2):249–260. doi: 10.1128/jvi.14.2.249-260.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu N. H., Radonovich M. F., Thoren M. M., Salzman N. P. Selective degradation of newly synthesized nonmessenger simian virus 40 transcripts. J Virol. 1978 Nov;28(2):590–599. doi: 10.1128/jvi.28.2.590-599.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell A. J., Kamen R. Strand-specific transcription of polyoma virus DNA late during productive infection. J Mol Biol. 1977 Sep 15;115(2):237–242. doi: 10.1016/0022-2836(77)90099-7. [DOI] [PubMed] [Google Scholar]
- Greenberg J. R. High stability of messenger RNA in growing cultured cells. Nature. 1972 Nov 10;240(5376):102–104. doi: 10.1038/240102a0. [DOI] [PubMed] [Google Scholar]
- Kamen R., Lindstrom D. M., Shure H., Old R. W. Virus-specific RNA in cells productively infected or transformed by polyoma virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):187–198. doi: 10.1101/sqb.1974.039.01.025. [DOI] [PubMed] [Google Scholar]
- Kamen R., Shure H. Topography of polyoma virus messenger RNA molecules. Cell. 1976 Mar;7(3):361–371. doi: 10.1016/0092-8674(76)90165-3. [DOI] [PubMed] [Google Scholar]
- Legon S., Flavell A. J., Cowie A., Kamen R. Amplification in the leader sequence of late polyoma virus mRNAs. Cell. 1979 Feb;16(2):373–388. doi: 10.1016/0092-8674(79)90013-8. [DOI] [PubMed] [Google Scholar]
- Lengyel J. A., Penman S. Differential stability of cytoplasmic RNA in a Drosophila cell line. Dev Biol. 1977 Jun;57(2):243–253. doi: 10.1016/0012-1606(77)90212-3. [DOI] [PubMed] [Google Scholar]
- Lengyel J., Penman S. hnRNA size and processing as related to different DNA content in two dipterans: Drosophila and Aedes. Cell. 1975 Jul;5(3):281–290. doi: 10.1016/0092-8674(75)90103-8. [DOI] [PubMed] [Google Scholar]
- Levis R., Penman S. The metabolism of poly (A)+ and poly(A)-hnRNA in cultured Drosophila cells studied with a rapid uridine pulse-chase. Cell. 1977 May;11(1):105–113. doi: 10.1016/0092-8674(77)90321-x. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Parish J. H., Kirby K. S. Reagents which reduce interactions between ribosomal RNA and rapidly labelled RNA from rat liver. Biochim Biophys Acta. 1966 Dec 21;129(3):554–562. doi: 10.1016/0005-2787(66)90070-0. [DOI] [PubMed] [Google Scholar]
- Penman S. RNA metabolism in the HeLa cell nucleus. J Mol Biol. 1966 May;17(1):117–130. doi: 10.1016/s0022-2836(66)80098-0. [DOI] [PubMed] [Google Scholar]
- Penman S., Vesco C., Penman M. Localization and kinetics of formation of nuclear heterodisperse RNA, cytoplasmic heterodisperse RNA and polyribosome-associated messenger RNA in HeLa cells. J Mol Biol. 1968 May 28;34(1):49–60. doi: 10.1016/0022-2836(68)90234-9. [DOI] [PubMed] [Google Scholar]
- Perry R. P., La Torre J., Kelley D. E., Greenberg J. R. On the lability of poly(A) sequences during extraction of messenger RNA from polyribosomes. Biochim Biophys Acta. 1972 Mar 14;262(2):220–226. doi: 10.1016/0005-2787(72)90236-5. [DOI] [PubMed] [Google Scholar]
- Philipson L., Pettersson U., Lindberg U., Tibbetts C., Vennström B., Persson T. RNA synthesis and processing in adenovirus-infected cells. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):447–456. doi: 10.1101/sqb.1974.039.01.057. [DOI] [PubMed] [Google Scholar]
- Piper P., Wardale J., Crew F. Splicing of the late mRNAs of polyoma virus does not occur in the cytoplasm of the infected cell. Nature. 1979 Dec 13;282(5740):686–691. doi: 10.1038/282686a0. [DOI] [PubMed] [Google Scholar]
- Rosenthal L. J. Isolation and characterization of poly(A)-containing polyoma "early" and "late" messenger RNAs. Nucleic Acids Res. 1976 Mar;3(3):661–676. doi: 10.1093/nar/3.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schochetman G., Perry R. P. Characterization of the messenger RNA released from L cell polyribosomes as a result of temperature shock. J Mol Biol. 1972 Feb 14;63(3):577–590. doi: 10.1016/0022-2836(72)90449-4. [DOI] [PubMed] [Google Scholar]
- Zuckermann M., Manor H., Parker J., Kamen R. Electron microscopic demonstration of the presence of amplified sequences at the 5'-ends of the polyoma virus late mRNAs. Nucleic Acids Res. 1980 Apr 11;8(7):1505–1519. doi: 10.1093/nar/8.7.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]


