Abstract
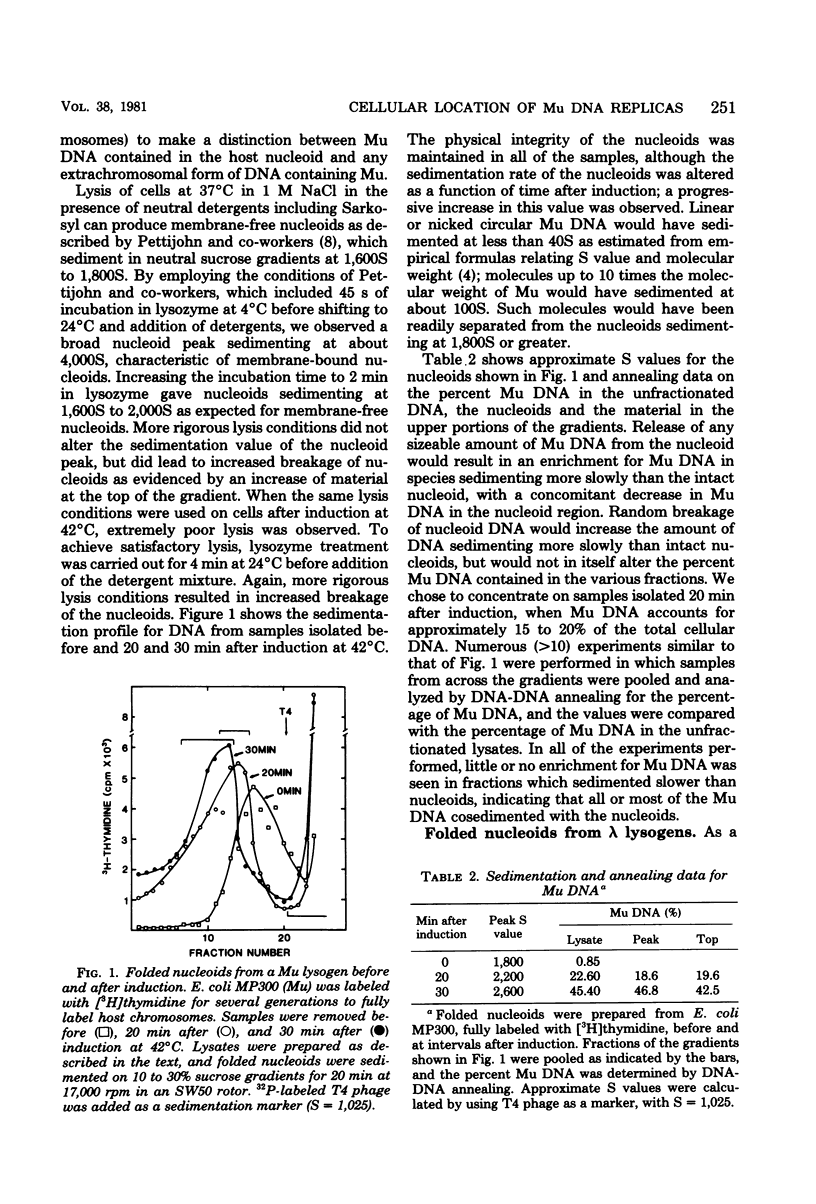
To ascertain the form and cellular location of the copies of bacteriophage Mu DNA synthesized during lytic development, DNA from an Escherichia coli lysogen was isolated at intervals after induction of the Mu prophage. Host chromosomes were isolated as intact, folded nucleoids, which could be digested with ribonuclease or heated in the presence of sodium dodecyl sulfate to yield intact, unfolded nucleoid DNA. Almost all of the Mu DNA in induced cells was associated with the nucleoids until shortly before cell lysis, even after unfolding of the nucleoid structure. We suggest that the replicas of Mu DNA are integrated into the host chromosomes, possibly by concerted replication-integration events, and are accumulated there until packaged shortly before cell lysis. Nucleoids also were isolated from induced lambda lysogens and from cells containing plasmid DNA. Most of the plasmid DNA sedimented independently of the unfolded nucleoid DNA, whereas 50% or more of the lambda DNA from induced lysogens cosedimented with unfolded nucleoid DNA. Possible explanations for the association of extrachromosomal DNA with nucleoid DNA are discussed.
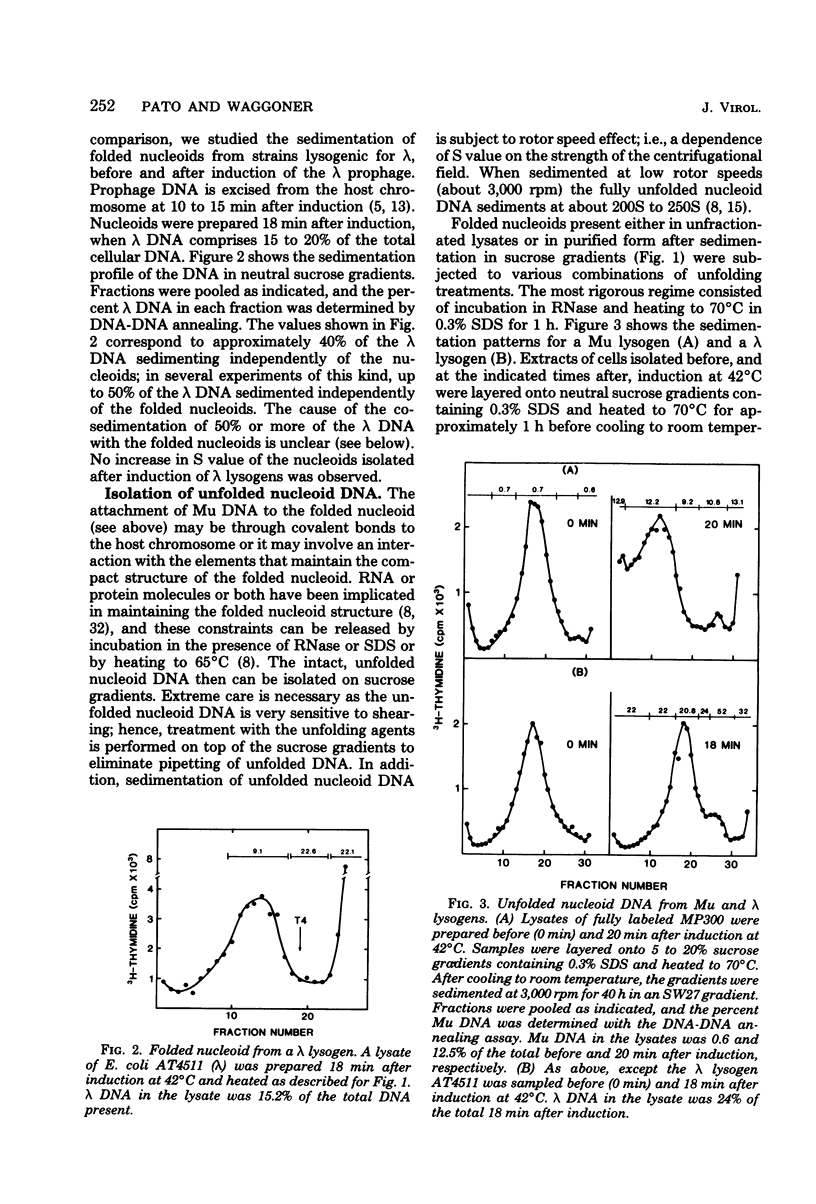
Full text
PDF

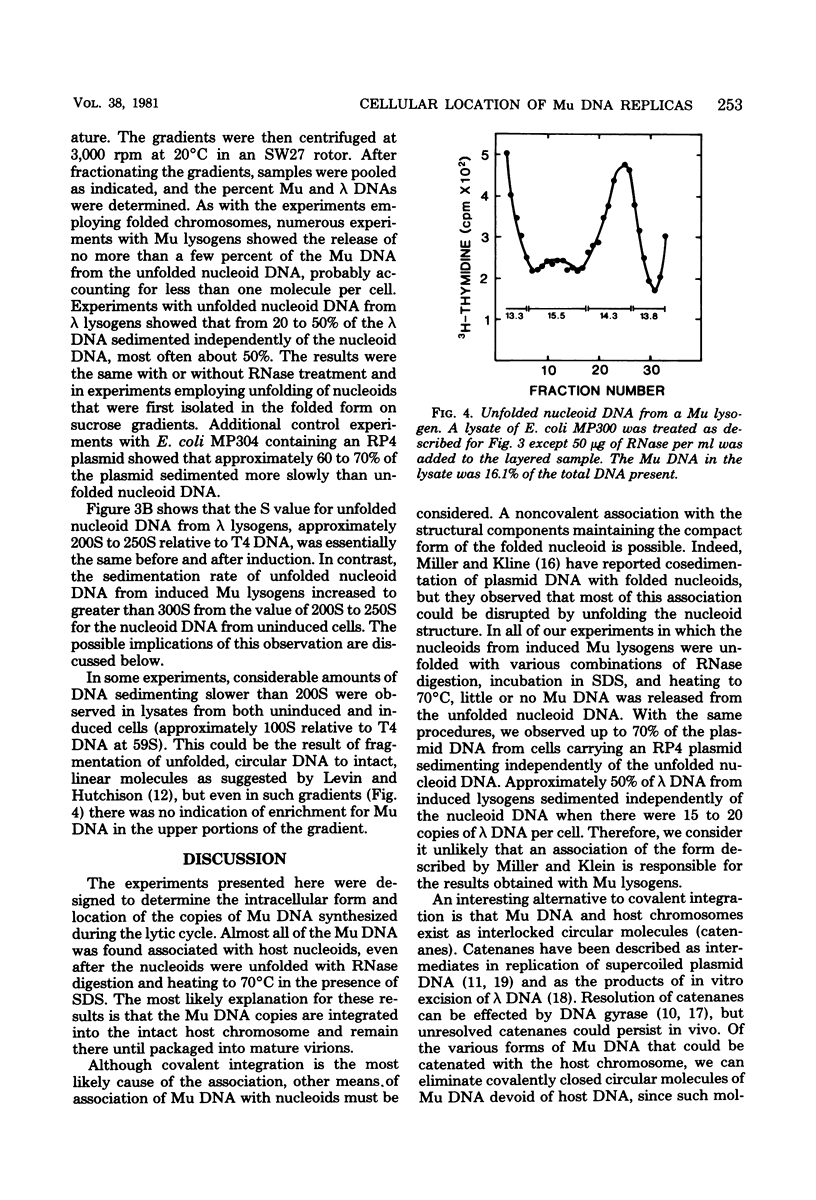


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bialy H., Waggoner B. T., Pato M. L. Fate of plasmids containing Mu DNA: chromosome association and mobilization. Mol Gen Genet. 1980;180(2):377–383. doi: 10.1007/BF00425851. [DOI] [PubMed] [Google Scholar]
- Chaconas G., Harshey R. M., Bukhari A. I. Association of Mu-containing plasmids with the Escherichia coli chromosome upon prophage induction. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1778–1782. doi: 10.1073/pnas.77.4.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifelder D., Kirschner I., Goldstein R., Baran N. Physical study of prophage excision and curing of lambda prophage from lysogenic Escherichia coli. J Mol Biol. 1973 Mar 15;74(4):703–720. doi: 10.1016/0022-2836(73)90058-2. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Sherratt D. J. Sequence analysis at IS1 insertion sites: models for transposition. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1257–1261. doi: 10.1101/sqb.1979.043.01.142. [DOI] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht R. M., Stimpson D., Pettijohn D. Sedimentation properties of the bacterial chromosome as an isolated nucleoid and as an unfolded DNA fiber. Chromosomal DNA folding measured by rotor speed effects. J Mol Biol. 1977 Apr 15;111(3):257–277. doi: 10.1016/s0022-2836(77)80051-x. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Cozzarelli N. R. Formation and resolution of DNA catenanes by DNA gyrase. Cell. 1980 May;20(1):245–254. doi: 10.1016/0092-8674(80)90252-4. [DOI] [PubMed] [Google Scholar]
- Kupersztoch Y. M., Helinski D. R. A catenated DNA molecule as an intermediate in the replication of the resistance transfer factor R6K in Escherichia coli. Biochem Biophys Res Commun. 1973 Oct 15;54(4):1451–1459. doi: 10.1016/0006-291x(73)91149-2. [DOI] [PubMed] [Google Scholar]
- Levin D., Hutchinson F. Neutral sucrose sedimentation of very large DNA from Bacillus subtilis. I. Effect of random double-strand breaks and centrifuge speed on sedimentation. J Mol Biol. 1973 Apr 15;75(3):455–478. doi: 10.1016/0022-2836(73)90454-3. [DOI] [PubMed] [Google Scholar]
- Ljungquist E., Bukhari A. I. Behavior of bacteriophage Mu DNA upon infecton of Escherichia coli cells. J Mol Biol. 1979 Sep 25;133(3):339–357. doi: 10.1016/0022-2836(79)90397-8. [DOI] [PubMed] [Google Scholar]
- Ljungquist E., Bukhari A. I. State of prophage Mu DNA upon induction. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3143–3147. doi: 10.1073/pnas.74.8.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydersen B. K., Pettijohn D. E. Interactions stabilizing DNA tertiary structure in the Escherichia coli chromosome investigated with ionizing radiation. Chromosoma. 1977 Jul 8;62(3):199–215. doi: 10.1007/BF00286044. [DOI] [PubMed] [Google Scholar]
- Miller J. R., Kline B. C. Biochemical characterization of nonintegrated plasmid-folded chromosome complexes: sex factor F and the Escherichia coli nucleoid. J Bacteriol. 1979 Feb;137(2):885–890. doi: 10.1128/jb.137.2.885-890.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Fisher L. M., O'Dea M. H., Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Smith K., Sheehy R. J., Murphy E. A catenated intermediate in plasmid replication. Biochem Biophys Res Commun. 1973 Oct 15;54(4):1460–1469. doi: 10.1016/0006-291x(73)91150-9. [DOI] [PubMed] [Google Scholar]
- Razzaki T., Bukhari A. I. Events following prophage Mu induction. J Bacteriol. 1975 May;122(2):437–442. doi: 10.1128/jb.122.2.437-442.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder W., Bade E. G., Delius H. Participation of Escherichia coli DNA in the replication of temperate bacteriophage Mu1. Virology. 1974 Aug;60(2):534–542. doi: 10.1016/0042-6822(74)90347-x. [DOI] [PubMed] [Google Scholar]
- Schröder W., van de Putte P. Genetic study of prophage excision with a temperature inducible mutant of Mu-1. Mol Gen Genet. 1974 May 21;130(2):99–104. doi: 10.1007/BF00269081. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snustad D. P., Parson K. A., Warner H. R., Tutas D. J., Wehner J. M., Koerner J. F. Mutants of bacteriophage T4 deficient in the ability to induce nuclear disruption. II. Physiological state of the host nucleoid in infected cells. J Mol Biol. 1974 Nov 15;89(4):675–687. doi: 10.1016/0022-2836(74)90044-8. [DOI] [PubMed] [Google Scholar]
- Van Brunt J., Waggoner B. T., Pato M. L. Re-examination of F plasmid replication in a dnaC mutant of Escherichia coli. Mol Gen Genet. 1977 Feb 15;150(3):285–292. doi: 10.1007/BF00268127. [DOI] [PubMed] [Google Scholar]
- Waggoner B. T., González N. S., Taylor A. L. Isolation of heterogeneous circular DNA from induced lysogens of bacteriophage Mu-1. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1255–1259. doi: 10.1073/pnas.71.4.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner B. T., Pato M. L. Early events in the replication of Mu prophage DNA. J Virol. 1978 Sep;27(3):587–594. doi: 10.1128/jvi.27.3.587-594.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijffelman C., Lotterman B. Kinetics of Mu DNA synthesis. Mol Gen Genet. 1977 Mar 7;151(2):169–174. doi: 10.1007/BF00338691. [DOI] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]


