Abstract
The recent crystal structure of complement protein component C2a reveals an interface between its VWA and serine protease domains that could not exist in the zymogen C2. The implied change in VWA domain conformation between C2 and C2a differs from that described for other VWA domains, including the I domains in integrins. Here, the remarkable diversity in both conformational regulation and ligand binding among VWA domains that function in complement, hemostasis, cell adhesion, anthrax toxin binding, vesicle transport, DNA break repair, and RNA quality control is reviewed. Finally, implications for metastability of complement convertases are discussed.
“Complement” is a system of over 30 plasma proteins that protect us from pathogens by sensitizing them for phagocytosis or directly lysing them. Complement fixation is initiated by three different pathways that all converge on activation of the component C3. Initial recognition of antibodies in the classical pathway and pathogen carbohydrates in the lectin pathway results in production of a C3 convertase consisting of the noncovalently associated C4b and C2a fragments (Figure 1). Initial recognition of distinctive features of pathogen surfaces in the alternative pathway results in formation of a distinct C3 convertase (not shown). This alternative C3 convertase consists of the C3b and factor Bb components, which are homologous to C4b and C2a, respectively. In the classical and lectin pathways, initial recognition is first amplified through the action of proteases C1s or MASP2, which cleave both C4 and C2 (Figure 1). In a second amplification step, C4bC2a proteolytically activates the pivotal C3 effector molecule (Fredslund et al., 2006; Janssen et al., 2005), i.e., acts as a C3 convertase (Figure 1). Cleavage is mediated by the serine protease domain of C2a, but an intact C4bC2a complex is required for C3 convertase activity. A large number of regulatory proteins in the complement system prevent amplification from spinning out of control.
Figure 1. Activation of C4bC2a in a Two-Stage Proteolytic Amplification Pathway that Results in the Cleavage of C3.
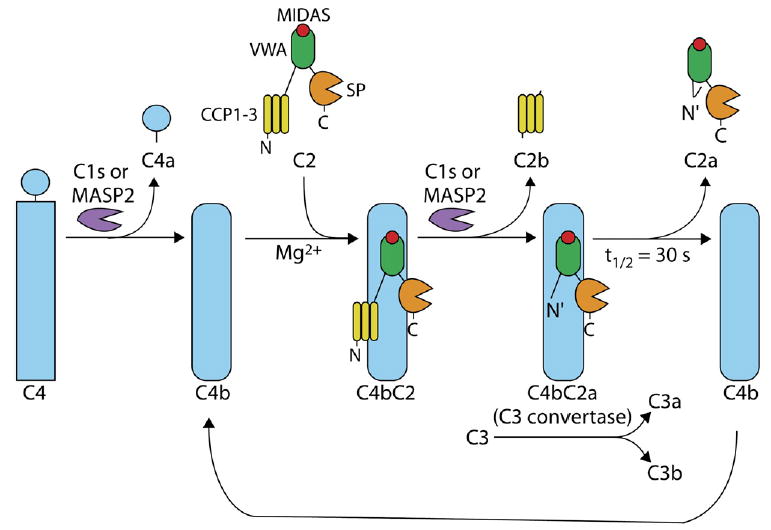
The first proteolytic amplification step in the classical and lectin complement pathways involves cleavage of C4 into C4b and C4a, Mg2+-dependent association of C2 with C4b, and cleavage of C4bC2 to C4bC2a, which results in formation of C3 convertase activity. The second amplification step is cleavage by C4bC2a of C3 to C3a and C3b. The buried N-terminal segment of C2a is shown with a bent line; burial could occur as shown here after release of C2a and help explain lack of ability of C2a to reassociate with C4b or could occur earlier, after cleavage of C4bC2 to C4bC2a. Adapted from Milder et al. (2006).
Another very interesting regulatory mechanism for keeping complement activation in check is metastability; i.e., the C3 convertases spontaneously decay by dissociation with a t1/2 of 30 s after their initial proteolytic activation (Kerr, 1980) (Figure 1). This limited stability ensures that the second proteolytic amplification step mediated by C3 convertases cannot proceed long in the absence of the first proteolytic amplification step mediated by C1s or MASP2 (Figure 1).
In a recent issue of Structure, Fin Milder, Piet Gros, and colleagues presented two elegant crystal structures of C2a (Milder et al., 2006). The C2a structure defines the conformation of C2a after it dissociates from C4b and is also relevant to the conformation that C2a assumes when associated with C4b in the C3 convertase (Figure 1). C2 has a three-lobed structure (Smith et al., 1984) consisting of three different types of domains: complement-control protein modules (CCPs), von Wille-brand factor A (VWA), and serine protease (SP) domains (Figure 1). Cleavage of C2 to generate C2a occurs between the CCP modules and the VWA domain (Figure 1). Strikingly, Milder et al. found that the newly created N-terminal segment of C2a is buried near the interface between the VWA and SP domains and therefore affects the relative orientation of these domains. Consequently, the two domains must differ in conformation between C2 and C2a (Milder et al., 2006). The following discussion of what is known about conformational change and ligand binding in structurally characterized VWA domains demonstrates remarkable diversity in structural regulation of ligand binding and the location of the ligand-binding site and provides a backdrop for understanding the role of the C2a VWA domain in the metastability of C4bC2a.
VWA Domains in Integrins Undergo Large Conformational Changes
VWA domains have a central β sheet with one antiparallel edge strand, amphipathic α helices that lie against each β sheet face, and often have a Mg2+ ion bound to the carboxy-terminal end of the β sheet, i.e., the “top” face of the domain. The antiparallel edge strand distinguishes VWA domains from the closely related Rossmann or nucleotide-binding domains, which bind nucleotide and have active enzyme sites in a position similar to the Mg2+ of the VWA domain. In VWA domains with a bound metal ion, it forms primary and secondary coordinations to residues in loops that constitute what is known as a metal-ion-dependent adhesion site (MIDAS) (Figures 1 and 2A). Of the three “MIDAS loops,” the first has an Asp-Xaa-Ser-Xaa-Ser (DXSXS) “MIDAS motif,” the second has a Thr, and the third has an Asp. The C and N termini of the VWA domain are close to one another at the “bottom” face, i.e., the face opposite the MIDAS site. In integrins, the VWA domains are inserted within the hybrid domain of the β subunit and, in some integrins, within the β-propeller domain of the α subunit and are therefore referred to as inserted (I) domains (Figures 2B and 2C). Both integrin α and β subunit I domains have open and closed conformations; α I domains additionally show an intermediate conformation (Luo et al., 2006). In the transition from closed to open, MIDAS loops change conformation, and the coordination with the Mg2+ ion is altered as an Asp moves out of and a Thr moves into the inner coordination shell, increasing the electrophilicity of the metal ion. Furthermore, in a linked rearrangement, the C-terminal α helix (α7) moves axially 7 Å toward the C terminus, i.e., “downward” (Figure 2B). Mutations designed to stabilize one or the other of these conformations, including introduction of disulfide bonds, demonstrate that the open conformation has much higher affinity for ligand than the closed conformation, in agreement with crystal studies that show that binding of a ligand, or binding to a pseudoligand in a lattice contact, stabilizes the open conformation (Luo et al., 2006). In the open conformation, the Mg2+ lacks any direct coordinations to negatively charged MIDAS residues. Therefore, binding to an acidic residue in the ligand is favored, and furthermore, the complementarity of other MIDAS loops for the ligand is also increased.
Figure 2. Representative Proteins Containing VWA Domains and Their Ligands.
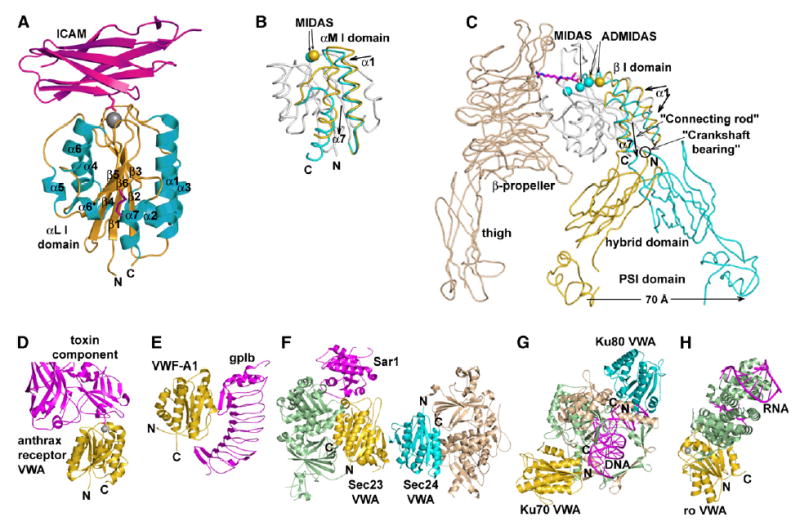
For orientation, the N and C termini of each VWA/I domain are marked. (A) VWA/I domain of the integrin αL subunit bound to domain 1 of ICAM-3 (PDB ID 1T0P). (B) Allostery in the integrin αM subunit I domain (PDB ID 1JLM and 1IDO). (C) Allostery in the integrin β3 subunit. The structures are composites of PDB ID 1U8C and 1TY6. A portion of a ligand is shown in magenta bound to the high-affinity conformation, for which the MIDAS Mg2+ and two adjacent Ca2+ ions are shown. In (B) and (C), regions that move in allostery are shown in gold (low affinity, unliganded state) and cyan (high affinity, liganded state), and nonmoving regions are in white (α or β I domains) or wheat (αIIb/αV subunit). See Luo et al. (2006) for references to structures in (A)–(C). (D) Anthrax receptor VWA domain bound to a toxin component, only a portion of which is shown. PDB ID 1T6B (Lacy et al., 2004). (E) The VWF-A1 domain bound to platelet glycoprotein gpIb, PDB ID 1M10 (Huizinga et al., 2002). (F) The sec23, sec24, and Sar1 GTPase complex. Composite of PDB ID 1M2O and 1M2V (Bi et al., 2002). (G) Complex of Ku70, Ku80, and DNA. PDB ID 1JEY (Walker et al., 2001). (H) Complex of Ro 60 kDa protein and RNA. PDB ID 1YVP (Stein et al., 2005).
In integrin VWA/I domains, the movement of the C-terminal α7 helix serves to convey allostery from the ligand binding face at the MIDAS to the opposite end of the domain, where it contacts neighboring domains (Figures 2B and 2C). In integrin β subunit I domains, the change in affinity at the MIDAS is linked to a piston and connecting-rod-like movement of the α7 helix connection to the hybrid domain and to pivoting about the other, crankshaft bearing-like connection between the I and hybrid domains (Figure 2C). This causes the hybrid domain to swing 60° and the “knees” of the integrin α and β subunits to separate by 70 Å.
Diverse Interactions between VWA Domains and Bound Ligands
There are similarities as well as differences in ligand binding among structurally characterized VWA domains. Two cell-surface molecules that contain an N-terminal VWA domain and function as receptors for laminin and collagen are co-opted as receptors for anthrax toxins. In the complex between the VWA domain and a toxin component, the VWA domain MIDAS Mg2+ coordinates an Asp on the toxin component (Santelli et al., 2004) (Figure 2D). The toxin-bound VWA I domain is in the open conformation. However, in two uncomplexed crystal structures, the anthrax toxin receptor VWA domain is also in the open conformation and binds to either a Glu of a neighboring VWA domain in the crystal lattice or to an acetate ion, i.e., to pseudoligands (Lacy et al., 2004). The anthrax toxin receptor appears to be more stable in the open conformation than integrin I domains, consistent with its high (200 pM) affinity.
von Willebrand factor (VWF) contains over 2,000 amino acid residues, including three VWA domains, and is polymerized through N- and C-terminal disulfide linkages into multimers that range in size up to 50,000,000 Da. High shear stress as found at sites of arterial bleeding activates VWF for binding to platelet glycoprotein Ib (gpIb) through the VWF-A1 domain. This domain lacks a MIDAS, owing to the lack of several of the residues required for metal-ion binding. Furthermore, the extensive binding site for gpIb is not on the top face, where the MIDAS is present in other VWA domains, but instead extends along the side bearing β strand 3 from the top face to the bottom face of the A1 domain (Figure 2E). No conformational change similar to that seen in integrins occurs in the VWA-A1 domain between crystal structures of the ligand-bound and free forms. However, changes are seen that are localized to the bottom face of the VWA domain. The conformations differ of the C- and N-terminal segments that in intact VWF would connect to adjacent domains; interestingly, mutations that activate binding to gpIb map to the bottom face of the A1 domain and to these connecting segments (Dumas et al., 2004; Emsley et al., 1998; Huizinga et al., 2002). Changes in these inter-domain linkers and the orientation of the VWF-A1 domain with respect to neighboring domains may be important in exposing the gpIb binding site in arterial bleeding, in which high shear would elongate VWF multimers.
The VWF-A3 domain binds to collagen. This domain also lacks an intact MIDAS, despite presence of a DXSXS motif, and collagen appears to bind to a distinct site near the bottom face of the domain (Bienkowska et al., 1997; Romijn et al., 2001). This contrasts with the integrin α2 I domain, which binds collagen at its MIDAS (Emsley et al., 2000).
Sec23 and Sec24 are structurally homologous proteins (Bi et al., 2002). Together with the GTPase Sar1, they form a prebudding complex on COPII vesicles that bud from the endoplasmic reticulum and traffic to the Golgi. Sec23 binds and acts as the GTPase-activating protein for Sar1, and Sec24 binds cargo and SNAREs. Sec23 and Sec24 contain VWA domains that form the Sec23/Sec24 trunk-like interface (Figure 2F). Their VWA domains are inserted in β-barrel domains, and thus, if the C-terminal α helix underwent connecting-rod-like displacements, the intersubunit orientation would shift as in integrin β subunits. Sec24 has a MIDAS motif (DXSXS) and nearby Ser, Thr, and Asp residues that might constitute a metal-ion binding site; however, no divalent cations were present during crystallization. Sec23 lacks a MIDAS motif. In Sec23, the VWA domain contacts the Switch 2 region of the Sar1 GTPase. The question of whether conformational change occurs in Sec23 or Sec24 VWA domains must await further study.
Ku70 and Ku80 are structurally homologous subunits of a protein that encircles DNA near double-strand breaks and facilitates repair by nonhomologous end joining (Walker et al., 2001). VWA domains at the N terminus of each subunit connect to β-barrel domains (Figure 2G). The VWA domains lack MIDAS motifs. The VWA domains interact only peripherally on their bottom faces with the bound DNA. Whether the VWA domains interact with other proteins involved in repair or undergo conformational change remains unknown.
The Ro 60 kDa autoantigen is a protein that binds mis-folded RNAs. It contains HEAT repeats that form an elliptical toroid that is closed at one end by a C-terminal VWA domain (Stein et al., 2005) (Figure 2H). RNA is bound over one face of the toroid and also in its central cavity. The VWA domain has a MIDAS in an open conformation. Crystals in Mg acetate have Mg2+ at the MIDAS and a liganded acetate ion. In the absence of Mg2+, a water molecule is found at the MIDAS, which retains the open conformation. It has been speculated that the VWA domain might act as a platform to recruit other factors involved in RNA quality control.
New Perspectives on VWA Domains
The first VWA domain structure was that of the integrin α I domain, which early on, was shown to transit between open and closed conformations (reviewed in Luo et al. [2006]). It seemed that conformational change might be paradigmatic for VWA domains. In support of this idea, GTPases and G protein α subunits, which also undergo allostery, have the closely related Rossmann fold topology, with a parallel instead of antiparallel edge strand. These G proteins bind Mg2+ in a site that resembles the MIDAS and coordinate the γ-phosphate of GTP similarly to the acidic residue of an integrin ligand. Furthermore, the presence of the γ-phosphate ligand in GTP or its absence in GDP control G protein allostery by modulating the structure of the Switch 1 and Switch 2 regions, although these bear no topological similarity to the α7 helix of the VWA domain.
The structures reviewed above show that generalizations about VWA domains can only be made with caution. Thus far, integrin α and β I domains are the only examples where conformational change at the MIDAS and piston or connecting rod-like movements of the α7 helix have been seen. Other structures with a MIDAS metal ion, the anthrax receptor, Ro, and as reviewed below, C2a and factor B, all have the MIDAS in the open conformation, and an alternative closed conformation has not yet been visualized. Other VWA domains, including the A1 and A3 domains of VWF, lack a MIDAS and bind ligand on a different face. Different types of conformational movements, in interdomain linkers on the bottom of these domains, may regulate exposure of the ligand-binding site on one side of the domain. However, thus far, only integrin β I domains reveal how a conformational change in a VWA domain relates to altered domain-domain interactions (Figure 2C).
The structure of C2a offers a refreshing new view of regulated interactions between VWA domains and neighboring domains (Milder et al., 2006) (Figure 3). Previous structures of complement factor B, which is 39% identical to C2 and has an analogous function in the alternative complement pathway, have revealed the isolated VWA and SP domains and the VWA and SP domains in tandem (Bhattacharya et al., 2004; Jing et al., 2000; Ponnuraj et al., 2004). Although a recombinant fragment of factor B containing the VWA and SP domains was used in crystallization, it was missing N-terminal residues present in the physiologic fragment Bb. In C2a, the analogous residues affect the VWA-SP interface.
Figure 3. Structure of C2a.
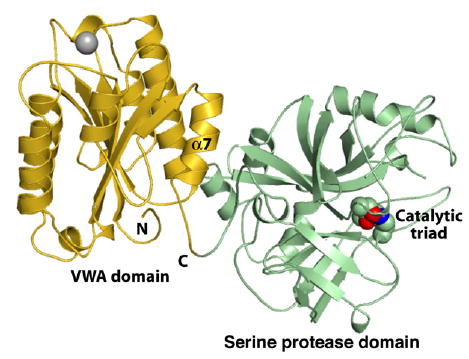
The N terminus created after cleavage of C2 to C2a nestles in a hydrophobic pocket on the bottom of the domain and affects the interface between the VWA and serine protease domains (Milder et al., 2006). Catalytic triad side-chain atoms are shown as spheres. Triad side chains are in the correct position for catalytic activity, but the oxyanion hole is incompletely formed and requires a flip of one peptide backbone moiety for activation. It is thought that this flip might be substrate induced. PDB ID 2I6Q (Milder et al., 2006).
The C2a structure (Milder et al., 2006) not only answers an important biological question about the structure of a crucial component in the complement pathway but also, like many important advances, allows longstanding, yet unsolved questions to be posed more precisely. How are the structures of C2, C4bC2, C4bC2a, and C2a related, in which the tandem unit containing the VWA and SP domains could theoretically exist in as many as four distinct conformational states? What is the basis for the metastability of C4bC2a, and the inability of C2a, once dissociated from the C4bC2a complex, to reassociate with C4b?
Remarkably, it is likely that in all four states, i.e., in C2, C4bC2, C4bC2a, and C2a, the VWA domain is predominantly in the open conformation. While there are thus far no structures for C2, C4bC2, or C4bC2a, the initial binding of C2 to C4 requires Mg2+. Furthermore, loop exchanges between C2 and factor B show that binding specificity requires the VWA domain MIDAS loops (Tuckwell et al., 1997). Moreover, in the DCSQS sequence of the C2 MIDAS motif, mutation or chemical modification of the cysteine and mutation of the glutamine affect the dissociation kinetics of the C4bC2a complex (Horiuchi et al., 1991). The isolated factor B VWA domain crystallizes in the open conformation with a ligand-mimetic crystal lattice contact (Bhattacharya et al., 2004). In a Bb-like fragment and C2a, the VWA domain can crystallize in the open conformation without a bound pseudoligand (Milder et al., 2006; Ponnuraj et al., 2004). The VWA domain MIDAS is clearly required for binding of C2 to C4b in C4bC2 and for binding of C2a to C4b in the C4bC2a complex. One therefore expects that these will have an open conformation of the VWA domain. In contrast, it remains a mystery why C2a retains an open MIDAS conformation, since once released, it cannot rebind C4b. However, the affinity for C4b of the VWA domain in different complexes and fragments could differ, if population of the postulated closed conformation (Bhattacharya et al., 2004) differed significantly. For example, VWA domains with open/closed population ratios of 90/10 and 99.99/0.01 would differ in affinity by approximately 1000-fold.
As described above, the metastability of C3 convertases is one of several mechanisms for ensuring that amplification in the complement pathway does not spin out of control. When C2 in the C4bC2 complex is cleaved to C2a, the C4bC2a complex becomes a metastable C3 convertase (Figure 1). C2 cleaved in solution to C2a, or C2a that has dissociated from C4b, when mixed with C4b, cannot bind to C4b or reconstitute C3 convertase activity (Kerr, 1980). There is evidence that the C2b fragment has affinity for C4b, and since both C2a and C2b have binding sites for C4b (Kerr, 1980), their combination in C2 should yield higher affinity. The alternative explanation for lack of reassociation would be an essentially irreversible conformational change in C2a. The decay of C3 convertase has been shown to reflect the dissociation of C2a from C4b, with a koff of 0.03 s−1 (Kerr, 1980). A koff in this range is typical for interactions of moderate to high affinity, and the extremely low kon that must be paired with this koff to give undetectable binding raises the possibility that a conformational change may occur in C2a after it dissociates from C4b that prevents it from rebinding (Figure 1).
Surprisingly, the structure of C2a shows that after proteolytic removal of the C2b fragment, the newly created N-terminal segment nestles in a cavity at the bottom of the VWA domain and affects the interface with the SP domain (Milder et al., 2006) (Figure 3). This segment must be accessible for proteolyic cleavage in C2 and C4bC2, and, therefore, its conformation must change in C2a. Milder et al. also point out that the nestling of the N-terminal segment also affects the conformation of the C-terminal α7 helix of the VWA domain and that it is in an intermediate conformational state similar to that seen previously for the integrin αL I domain (Luo et al., 2006). Furthermore, the binding of the N-terminal segment to a pocket on the bottom face of the VWA domain demonstrates a novel conformational mechanism for regulating the orientation of VWA domains with respect to neighboring domains. Perhaps the closest analogy among structurally characterized VWA domains would be the VWF-A1 domain, where mutations on its bottom face demonstrate the importance of the bottom face in allosterically regulating ligand binding to the side of the VWA domain (Dumas et al., 2004; Huizinga et al., 2002).
Questions for the Future
Because of metastability, a crystal structure of C4bC2a would be most challenging. However, the work of Milder et al. whets the appetite for structures of C2, C4bC2, and C4bC2a that would answer further remaining questions on the putative interaction of the C2a VWF domain MI-DAS with C4b, the basis for the catalytic activity and metastability of C3 convertases, and how the conformations of the VWA domain and its N-terminal segment differ from C2, C4bC2, C4bC2a, and C2a. For other classes of VWA domains, future structures should reveal whether the paradigm of shape shifting between open and closed conformations can be extended beyond integrins, whether other VWA domains with intact MIDAS bind ligands at this site, and further diversity in how conformational movements at the bottom of the VWA domain regulate interactions with adjacent N- and C-terminally connected domains.
Acknowledgments
The author’s work is supported by National Institutes of Health grants CA31799, AI72765, and HL48675 and by the Latham Family Professorship. I thank Dr. Bing-Hao Luo for help with Figures 2A–2C.
References
- Bhattacharya AA, Lupher ML, Jr, Staunton DE, Liddington RC. Crystal structure of the A domain from complement factor B reveals an integrin-like open conformation. Structure. 2004;12:371–378. doi: 10.1016/j.str.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 prebudding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- Bienkowska J, Cruz M, Atiemo A, Handin R, Liddington R. The von Willebrand factor A3 domain does not contain a metal ion-dependent adhesion site motif. J Biol Chem. 1997;272:25162–25167. doi: 10.1074/jbc.272.40.25162. [DOI] [PubMed] [Google Scholar]
- Dumas JJ, Kumar R, McDonagh T, Sullivan F, Stahl ML, Somers WS, Mosyak L. Crystal structure of the wild-type von Willebrand factor A1-glycoprotein Ibalpha complex reveals conformation differences with a complex bearing von Willebrand disease mutations. J Biol Chem. 2004;279:23327–23334. doi: 10.1074/jbc.M401659200. [DOI] [PubMed] [Google Scholar]
- Emsley J, Cruz M, Handin R, Liddington R. Crystal structure of the von Willebrand Factor A1 domain and implications for the binding of platelet glycoprotein Ib. J Biol Chem. 1998;273:10396–10401. doi: 10.1074/jbc.273.17.10396. [DOI] [PubMed] [Google Scholar]
- Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Structural basis of collagen recognition by integrin α2β1. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- Fredslund F, Jenner L, Husted LB, Nyborg J, Andersen GR, Sottrup-Jensen L. The structure of bovine complement component 3 reveals the basis for thioester function. J Mol Biol. 2006;361:115–127. doi: 10.1016/j.jmb.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Macon KJ, Engler JA, Volanakis JE. Site-directed mutagenesis of the region around Cys-241 of complement component C2. Evidence for a C4b binding site. J Immunol. 1991;147:584–589. [PubMed] [Google Scholar]
- Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P. Structures of glycoprotein Ibα and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Huizinga EG, Raaijmakers HC, Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- Jing H, Xu Y, Carson M, Moore D, Macon KJ, Volanakis JE, Narayana SV. New structural motifs on the chymotrypsin fold and their potential roles in complement factor B. EMBO J. 2000;19:164–173. doi: 10.1093/emboj/19.2.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MA. The human complement system: assembly of the classical pathway C3 convertase. Biochem J. 1980;189:173–181. doi: 10.1042/bj1890173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy DB, Wigelsworth DJ, Scobie HM, Young JAT, Collier RJ. Crystal structure of the von Willenbrand factor A domain of human capillary morphogenesis protein 2: an anthrax toxin receptor. Proc Natl Acad Sci USA. 2004;101:6367–6372. doi: 10.1073/pnas.0401506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B-H, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2006 doi: 10.1146/annurev.immunol.25.022106.141618. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milder FJ, Raaijmakers HCA, Vandeputte MDAA, Schouten A, Huizinga EG, Romijn RA, Hemrika W, Roos A, Daha MR, Gros P. Structure of complement component C2a: implications for convertase formation and substrate binding. Structure. 2006;14:1587–1597. doi: 10.1016/j.str.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Ponnuraj K, Xu Y, Macon K, Moore D, Volanakis JE, Narayana SV. Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol Cell. 2004;14:17–28. doi: 10.1016/s1097-2765(04)00160-1. [DOI] [PubMed] [Google Scholar]
- Romijn RA, Bouma B, Wuyster W, Gros P, Kroon J, Sixma JJ, Huizinga EG. Identification of the collagen-binding site of the von Willebrand factor A3-domain. J Biol Chem. 2001;276:9985–9991. doi: 10.1074/jbc.M006548200. [DOI] [PubMed] [Google Scholar]
- Santelli E, Bankston LA, Leppla SH, Liddington RC. Crystal structure of a complex between anthrax toxin and its host cell receptor. Nature. 2004;430:905–908. doi: 10.1038/nature02763. [DOI] [PubMed] [Google Scholar]
- Smith CA, Vogel CW, Müller-Eberhard HJ. MHC class III products: an electron microscopic study of the C3 convertases of human complement. J Exp Med. 1984;159:324–329. doi: 10.1084/jem.159.1.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–539. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckwell DS, Xu Y, Newham P, Humphries MJ, Volanakis JE. Surface loops adjacent to the cation-binding site of the complement factor B von Willebrand factor type A module determine C3b binding specificity. Biochemistry. 1997;36:6605–6613. doi: 10.1021/bi963155l. [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]


