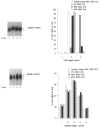
Abstract
While mtDNA polymorphisms at single base positions are common, the overwhelming majority of the mitochondrial genomes within a single individual are usually identical. When there is a point-mutation difference between a mother and her offspring, there may be a complete switching of mtDNA type within a single generation. It is generally assumed that there is a genetic bottleneck whereby a single or small number of founder mtDNA(s) populate the organism, but it is not known at which stages the restriction/amplification of mtDNA subtype(s) occur, and this uncertainty impedes antenatal diagnosis for mtDNA disorders. Length polymorphisms in homopolymeric tracts have been demonstrated in the large noncoding region of mtDNA. We have developed a new method, T-PCR (trimmed PCR), to quantitate heteroplasmy for two of these tracts (D310 and D16189). D310 variation is sufficient to indicate clonal origins of tissues and single oocytes. Tissues from normal individuals often possessed more than one length variant (heteroplasmy). However, there was no difference in the pattern of the length variants between somatic tissues in any control individual when bulk samples were taken. Oocytes from normal women undergoing in vitro fertilization were frequently heteroplasmic for length variants, and in two cases the modal length of the D310 tract differed in individual oocytes from the same woman. These data suggest that a restriction/amplification event, which we attribute to clonal expansion of founder mtDNA(s), has occurred by the time oocytes are mature, although further segregation may occur at a later stage. In contrast to controls, the length distribution of the D310 tract varied between tissues in a patient with heteroplasmic mtDNA rearrangements, suggesting that these mutants influence segregation. These findings have important implications for the genetic counselling of patients with pathogenic mtDNA mutations.
Full text
PDF


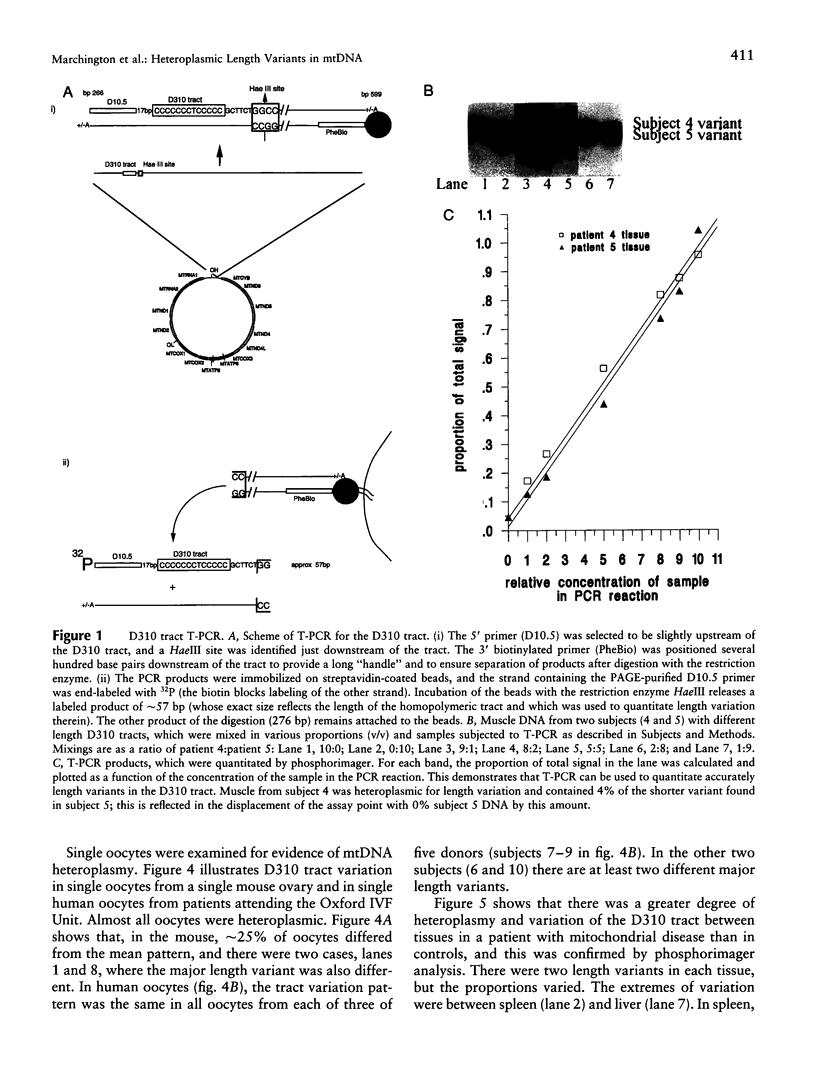
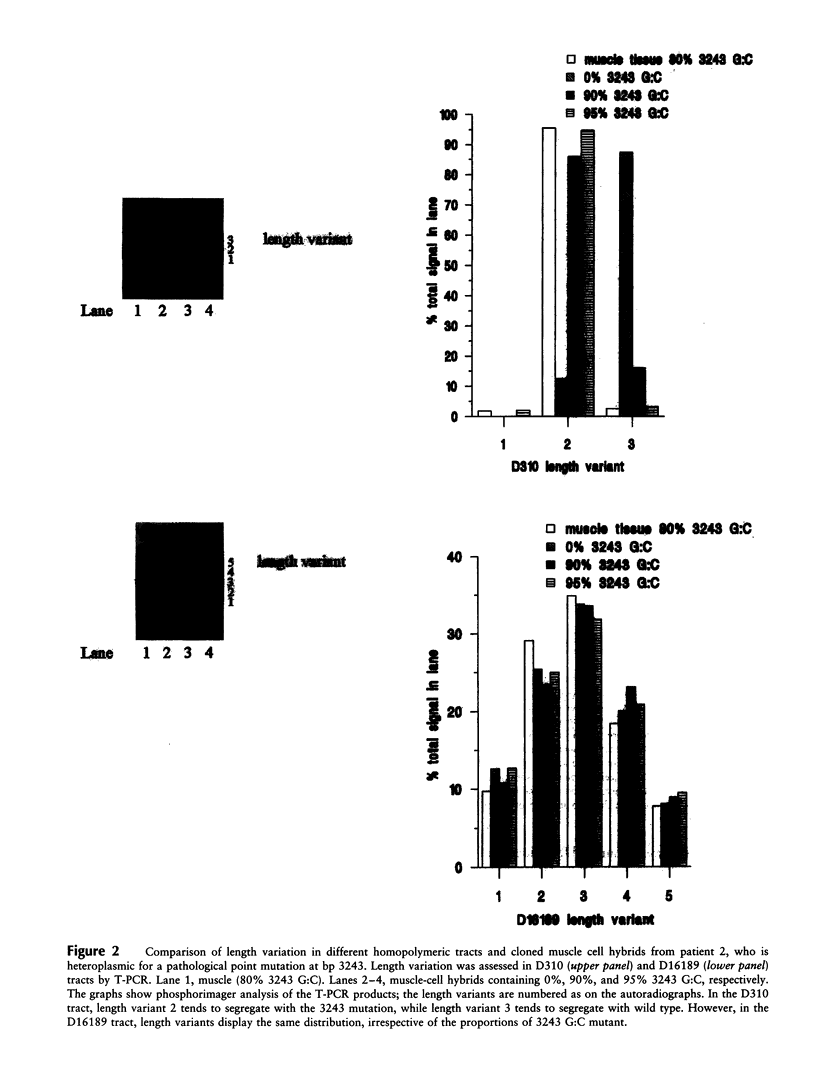
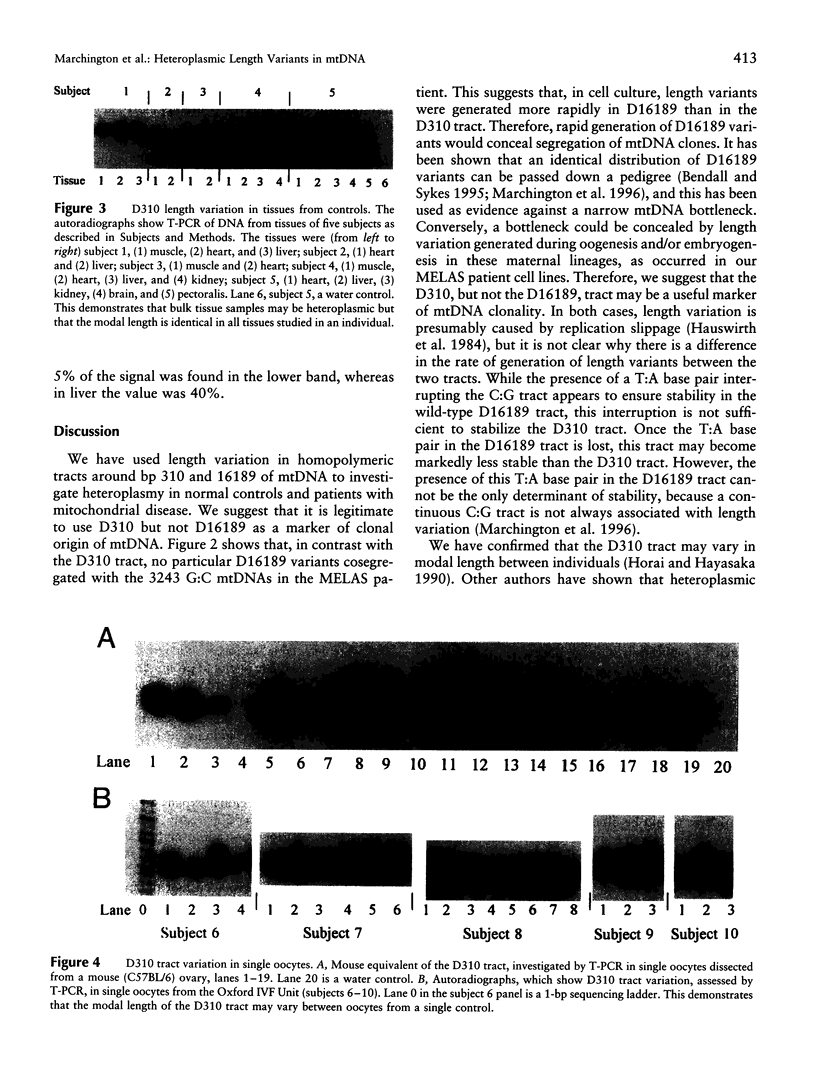



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashley M. V., Laipis P. J., Hauswirth W. W. Rapid segregation of heteroplasmic bovine mitochondria. Nucleic Acids Res. 1989 Sep 25;17(18):7325–7331. doi: 10.1093/nar/17.18.7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall K. E., Sykes B. C. Length heteroplasmy in the first hypervariable segment of the human mtDNA control region. Am J Hum Genet. 1995 Aug;57(2):248–256. [PMC free article] [PubMed] [Google Scholar]
- Dunbar D. R., Moonie P. A., Jacobs H. T., Holt I. J. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc Natl Acad Sci U S A. 1995 Jul 3;92(14):6562–6566. doi: 10.1073/pnas.92.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth W. W., Clayton D. A. Length heterogeneity of a conserved displacement-loop sequence in human mitochondrial DNA. Nucleic Acids Res. 1985 Nov 25;13(22):8093–8104. doi: 10.1093/nar/13.22.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth W. W., Van de Walle M. J., Laipis P. J., Olivo P. D. Heterogeneous mitochondrial DNA D-loop sequences in bovine tissue. Cell. 1984 Jul;37(3):1001–1007. doi: 10.1016/0092-8674(84)90434-3. [DOI] [PubMed] [Google Scholar]
- Horai S., Hayasaka K. Intraspecific nucleotide sequence differences in the major noncoding region of human mitochondrial DNA. Am J Hum Genet. 1990 Apr;46(4):828–842. [PMC free article] [PubMed] [Google Scholar]
- Howell N., Halvorson S., Kubacka I., McCullough D. A., Bindoff L. A., Turnbull D. M. Mitochondrial gene segregation in mammals: is the bottleneck always narrow? Hum Genet. 1992 Sep-Oct;90(1-2):117–120. doi: 10.1007/BF00210753. [DOI] [PubMed] [Google Scholar]
- Jenuth J. P., Peterson A. C., Fu K., Shoubridge E. A. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet. 1996 Oct;14(2):146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- Koehler C. M., Lindberg G. L., Brown D. R., Beitz D. C., Freeman A. E., Mayfield J. E., Myers A. M. Replacement of bovine mitochondrial DNA by a sequence variant within one generation. Genetics. 1991 Sep;129(1):247–255. doi: 10.1093/genetics/129.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshon D., Zweifel S. G., Freeman-Cook L. L., Lorimer H. E., Brewer B. J., Fangman W. L. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell. 1995 Jun 16;81(6):947–955. doi: 10.1016/0092-8674(95)90014-4. [DOI] [PubMed] [Google Scholar]
- Marchington D. R., Poulton J., Sellar A., Holt I. J. Do sequence variants in the major non-coding region of the mitochondrial genome influence mitochondrial mutations associated with disease? Hum Mol Genet. 1996 Apr;5(4):473–479. doi: 10.1093/hmg/5.4.473. [DOI] [PubMed] [Google Scholar]
- Matthews P. M., Hopkin J., Brown R. M., Stephenson J. B., Hilton-Jones D., Brown G. K. Comparison of the relative levels of the 3243 (A-->G) mtDNA mutation in heteroplasmic adult and fetal tissues. J Med Genet. 1994 Jan;31(1):41–44. doi: 10.1136/jmg.31.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monckton D. G., Neumann R., Guram T., Fretwell N., Tamaki K., MacLeod A., Jeffreys A. J. Minisatellite mutation rate variation associated with a flanking DNA sequence polymorphism. Nat Genet. 1994 Oct;8(2):162–170. doi: 10.1038/ng1094-162. [DOI] [PubMed] [Google Scholar]
- Monnat R. J., Jr, Loeb L. A. Nucleotide sequence preservation of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1985 May;82(9):2895–2899. doi: 10.1073/pnas.82.9.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton J., Morten K. Noninvasive diagnosis of the MELAS syndrome from blood DNA. Ann Neurol. 1993 Jul;34(1):116–116. doi: 10.1002/ana.410340124. [DOI] [PubMed] [Google Scholar]
- Poulton J., O'Rahilly S., Morten K. J., Clark A. Mitochondrial DNA, diabetes and pancreatic pathology in Kearns-Sayre syndrome. Diabetologia. 1995 Jul;38(7):868–871. doi: 10.1007/s001250050366. [DOI] [PubMed] [Google Scholar]
- Solignac M., Génermont J., Monnerot M., Mounolou J. C. Drosophila Mitochondrial Genetics: Evolution of Heteroplasmy through Germ Line Cell Divisions. Genetics. 1987 Dec;117(4):687–696. doi: 10.1093/genetics/117.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda M., Chomyn A., Martinuzzi A., Hurko O., Attardi G. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyopathy. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11164–11168. doi: 10.1073/pnas.89.23.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]