Abstract
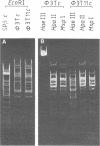
The resistance of Φ3T DNA to degradation by the restriction enzyme BsuR or its isoschizomer HaeIII is due to obligatory modification of such DNA. Biochemical and genetical experiments indicate that Φ3T codes for a methyltransferase, which methylates Φ3T DNA itself or heterologous DNA at target sites 5′-GG*CC.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bron S., Hörz W. Purification and properties of the Bsu endonuclease. Methods Enzymol. 1980;65(1):112–132. doi: 10.1016/s0076-6879(80)65017-4. [DOI] [PubMed] [Google Scholar]
- Cregg J. M., Ito J. A physical map of the genome of temperate phage phi 3T. Gene. 1979 Jul;6(3):199–219. doi: 10.1016/0378-1119(79)90058-1. [DOI] [PubMed] [Google Scholar]
- Cregg J. M., Nguyen A. H., Ito J. DNA modification induced during infection of Bacillus subtilis by phage phi 3T. Gene. 1980 Dec;12(1-2):17–24. doi: 10.1016/0378-1119(80)90011-6. [DOI] [PubMed] [Google Scholar]
- Duncan C. H., Wilson G. A., Young F. E. Mechanism of integrating foreign DNA during transformation of Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3664–3668. doi: 10.1073/pnas.75.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunthert U., Schweiger M., Stupp M., Doerfler W. DNA methylation in adenovirus, adenovirus-transformed cells, and host cells. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3923–3927. doi: 10.1073/pnas.73.11.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Pawlek B., Stutz J., Trautner T. A. Restriction and modification in Bacillus subtilis: inducibility of a DNA methylating activity in nonmodifying cells. J Virol. 1976 Oct;20(1):188–195. doi: 10.1128/jvi.20.1.188-195.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Storm K., Bald R. Restriction and modification in Bacillus subtilis. Localization of the methylated nucleotide in the BsuRI recognition sequence. Eur J Biochem. 1978 Oct 16;90(3):581–583. doi: 10.1111/j.1432-1033.1978.tb12638.x. [DOI] [PubMed] [Google Scholar]
- Günthert U., Stutz J., Klotz G. Restriction and modification in B. subtilis. The biochemical basis of modification against endo R. Bsu R restriction. Mol Gen Genet. 1975 Dec 30;142(3):185–191. doi: 10.1007/BF00425644. [DOI] [PubMed] [Google Scholar]
- Ito J., Roberts R. J. Unusual base sequence arrangement in phage phi 29 DNA. Gene. 1979 Jan;5(1):1–7. doi: 10.1016/0378-1119(79)90088-x. [DOI] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palefski S., Hemphill H. E., Kolenbrander P. E., Whiteley H. R. Dominance relationships in mixedly infected Bacillus subtilis. J Virol. 1972 Apr;9(4):594–601. doi: 10.1128/jvi.9.4.594-601.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N., Amann E., Tailor R., Günthert U., Scholz K., Trautner T. A. Unusual behaviour of SPO1 DNA with respect to restriction and modification enzymes recognizing the sequence 5'-G-G-C-C. Mol Gen Genet. 1980 Apr;178(1):229–231. doi: 10.1007/BF00267234. [DOI] [PubMed] [Google Scholar]
- Trautner T. A., Pawlek B., Bron S., Anagnostopoulos C. Restriction and modification in B. subtilis. Biological aspects. Mol Gen Genet. 1974;131(3):181–191. doi: 10.1007/BF00267958. [DOI] [PubMed] [Google Scholar]
- Trautner T. A., Pawlek B., Günthert U., Canosi U., Jentsch S., Freund M. Restriction and modification in Bacillus subtilis: identification of a gene in the temperate phage SP beta coding for a BsuR specific modification methyltransferase. Mol Gen Genet. 1980;180(2):361–367. doi: 10.1007/BF00425849. [DOI] [PubMed] [Google Scholar]
- Tucker R. G. Acquisition of thymidylate synthetase activity by a thymine-requiring mutant of Bacillus subtilis following infection by the temperate phage phi 3. J Gen Virol. 1969 Jun;4(4):489–504. doi: 10.1099/0022-1317-4-4-489. [DOI] [PubMed] [Google Scholar]
- Williams M. T., Young F. E. Temperate Bacillus subtilis bacteriophage phi 3T: chromosomal attachment site and comparison with temperate bacteriophages phi 105 and SPO2. J Virol. 1977 Feb;21(2):522–529. doi: 10.1128/jvi.21.2.522-529.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler S. A., Korman R. Z., Rosenthal R., Hemphill H. E. Bacillus subtilis bacteriophage SPbeta: localization of the prophage attachment site, and specialized transduction. J Bacteriol. 1977 Jan;129(1):556–558. doi: 10.1128/jb.129.1.556-558.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]