Abstract
Detection of linkage with a systematic genome scan in nuclear families including an affected sibling pair is an important initial step on the path to cloning susceptibility genes for complex genetic disorders, and it is desirable to optimize the efficiency of such studies. The aim is to maximize power while simultaneously minimizing the total number of genotypings and probability of type I error. One approach to increase efficiency, which has been investigated by other workers, is grid tightening: a sample is initially typed using a coarse grid of markers, and promising results are followed up by use of a finer grid. Another approach, not previously considered in detail in the context of an affected-sib-pair genome scan for linkage, is sample splitting: a portion of the sample is typed in the screening stage, and promising results are followed up in the whole sample. In the current study, we have used computer simulation to investigate the relative efficiency of two-stage strategies involving combinations of both grid tightening and sample splitting and found that the optimal strategy incorporates both approaches. In general, typing half the sample of affected pairs with a coarse grid of markers in the screening stage is an efficient strategy under a variety of conditions. If Hardy-Weinberg equilibrium holds, it is most efficient not to type parents in the screening stage. If Hardy-Weinberg equilibrium does not hold (e.g., because of stratification) failure to type parents in the first stage increases the amount of genotyping required, although the overall probability of type I error is not greatly increased, provided the parents are used in the final analysis.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown D. L., Gorin M. B., Weeks D. E. Efficient strategies for genomic searching using the affected-pedigree-member method of linkage analysis. Am J Hum Genet. 1994 Mar;54(3):544–552. [PMC free article] [PubMed] [Google Scholar]
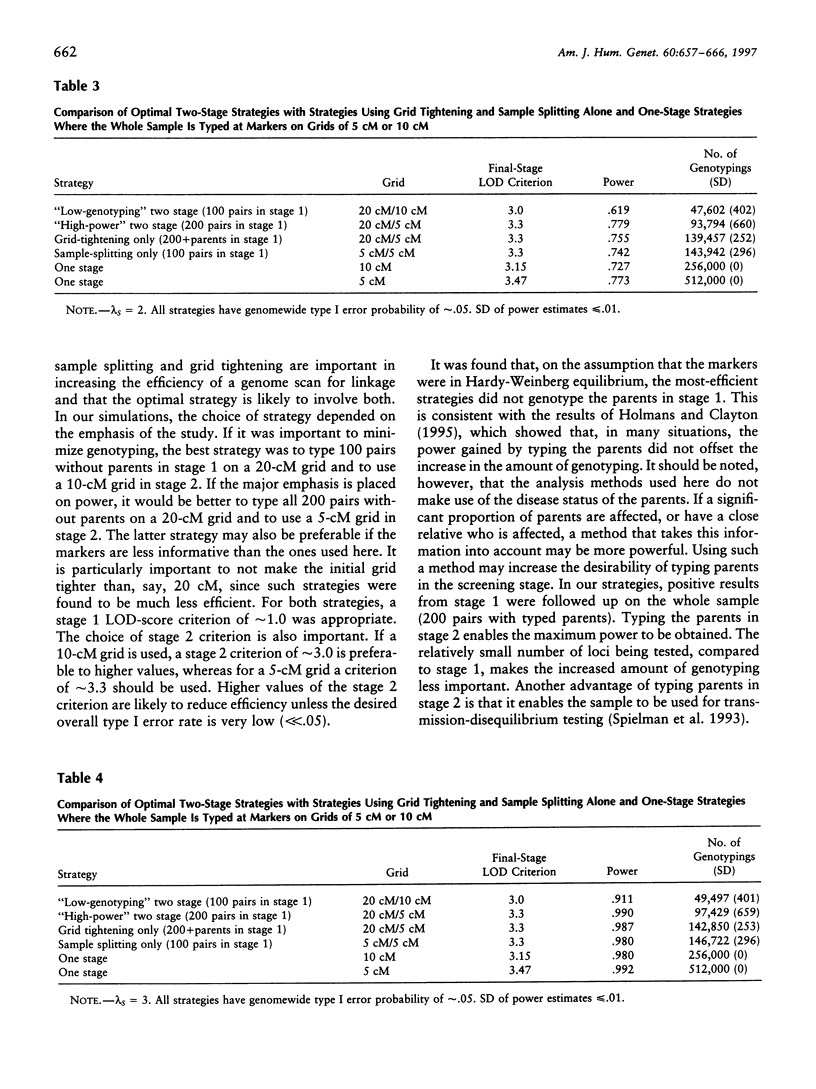
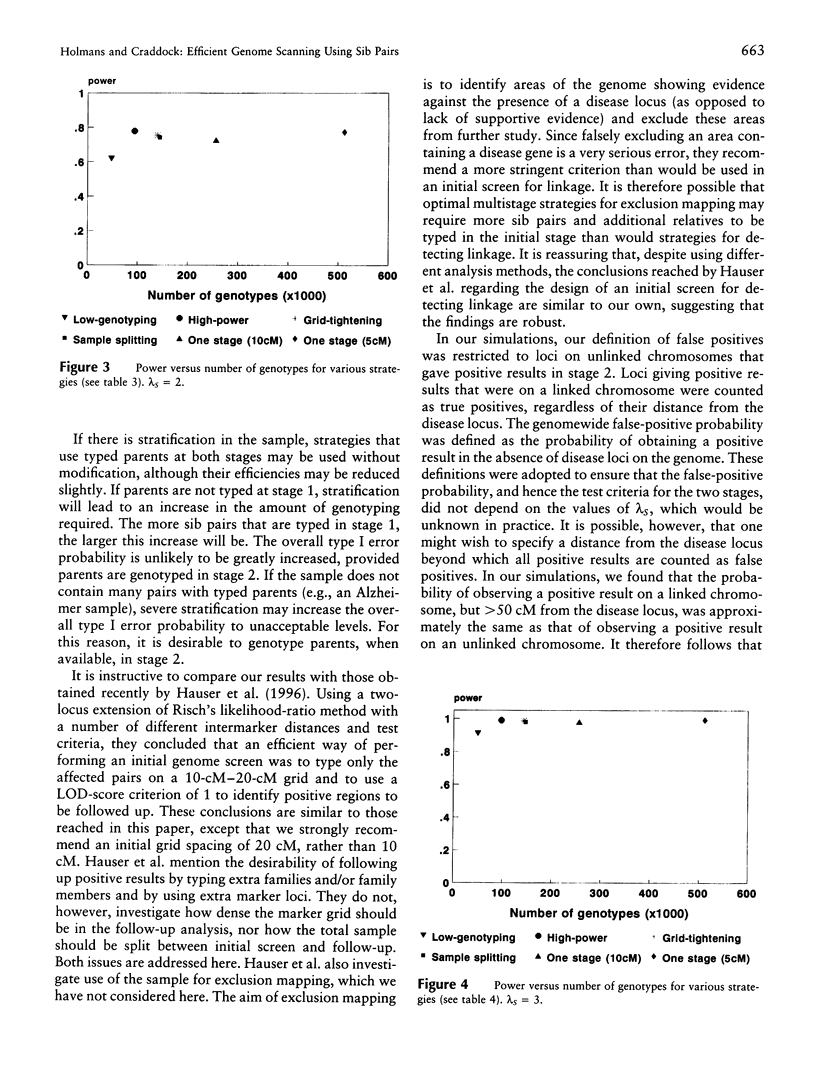
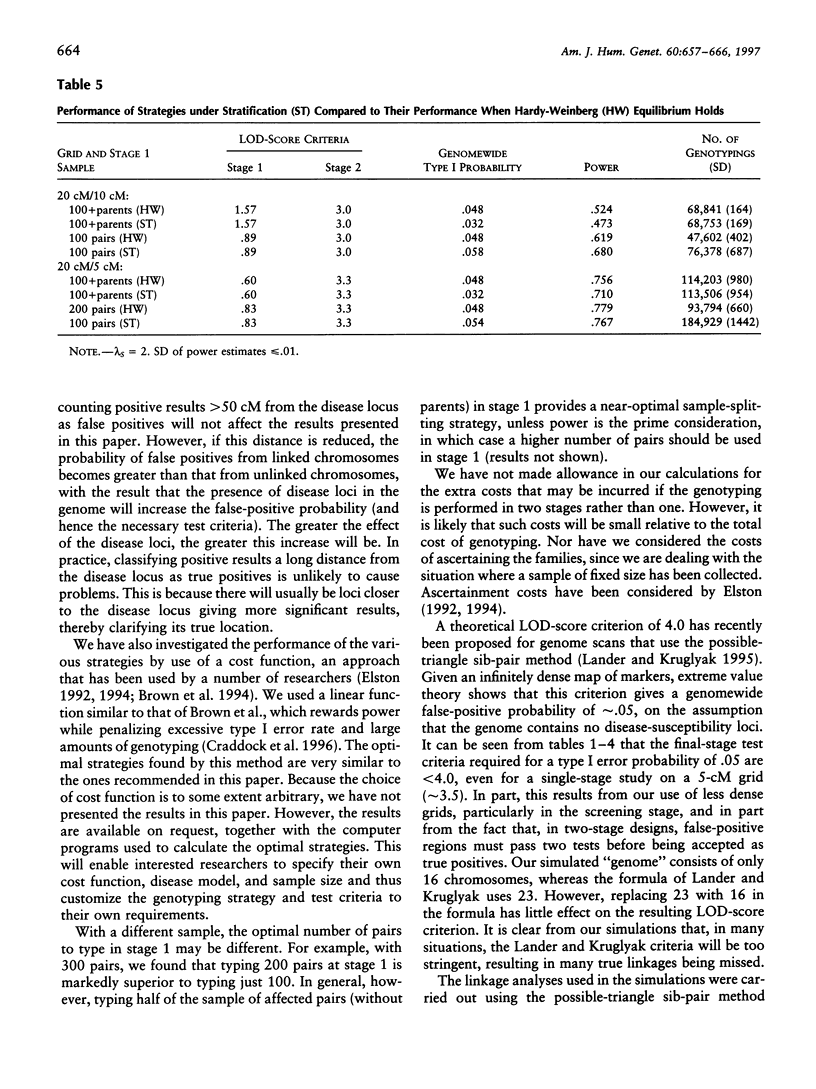
- Craddock N., Daniels J., Holmans P., Williams N., Owen M. J. Increasing the efficiency of genomic searches for linkage in complex disorders by DNA pooling of affected sib-pairs. Mol Psychiatry. 1996 Mar;1(1):59–64. [PubMed] [Google Scholar]
- Craddock N., Khodel V., Van Eerdewegh P., Reich T. Mathematical limits of multilocus models: the genetic transmission of bipolar disorder. Am J Hum Genet. 1995 Sep;57(3):690–702. [PMC free article] [PubMed] [Google Scholar]
- Craddock N., Owen M. J. Modern molecular genetic approaches to psychiatric disease. Br Med Bull. 1996 Jul;52(3):434–452. doi: 10.1093/oxfordjournals.bmb.a011558. [DOI] [PubMed] [Google Scholar]
- Davies J. L., Kawaguchi Y., Bennett S. T., Copeman J. B., Cordell H. J., Pritchard L. E., Reed P. W., Gough S. C., Jenkins S. C., Palmer S. M. A genome-wide search for human type 1 diabetes susceptibility genes. Nature. 1994 Sep 8;371(6493):130–136. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- Hauser E. R., Boehnke M., Guo S. W., Risch N. Affected-sib-pair interval mapping and exclusion for complex genetic traits: sampling considerations. Genet Epidemiol. 1996;13(2):117–137. doi: 10.1002/(SICI)1098-2272(1996)13:2<117::AID-GEPI1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Holmans P. Asymptotic properties of affected-sib-pair linkage analysis. Am J Hum Genet. 1993 Feb;52(2):362–374. [PMC free article] [PubMed] [Google Scholar]
- Holmans P., Clayton D. Efficiency of typing unaffected relatives in an affected-sib-pair linkage study with single-locus and multiple tightly linked markers. Am J Hum Genet. 1995 Nov;57(5):1221–1232. [PMC free article] [PubMed] [Google Scholar]
- Korczak J. F., Pugh E. W., Premkumar S., Guo X., Elston R. C., Bailey-Wilson J. E. Effects of marker information on sib-pair linkage analysis of a rare disease. Genet Epidemiol. 1995;12(6):625–630. doi: 10.1002/gepi.1370120617. [DOI] [PubMed] [Google Scholar]
- Kruglyak L., Lander E. S. Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet. 1995 Aug;57(2):439–454. [PMC free article] [PubMed] [Google Scholar]
- Lander E., Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995 Nov;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Nemesure B. B., He Q., Mendell N. Integration of linkage analyses and disease association studies. Genet Epidemiol. 1995;12(6):653–658. doi: 10.1002/gepi.1370120622. [DOI] [PubMed] [Google Scholar]
- Risch N. Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet. 1990 Feb;46(2):222–228. [PMC free article] [PubMed] [Google Scholar]
- Sham P. Sequential analysis and case-control candidate gene association studies: reply to Sobell et al. Am J Med Genet. 1994 Jun 15;54(2):154–157. doi: 10.1002/ajmg.1320540212. [DOI] [PubMed] [Google Scholar]
- Sobell J. L., Heston L. L., Sommer S. S. Novel association approach for determining the genetic predisposition to schizophrenia: case-control resource and testing of a candidate gene. Am J Med Genet. 1993 May 1;48(1):28–35. doi: 10.1002/ajmg.1320480108. [DOI] [PubMed] [Google Scholar]
- Spielman R. S., McGinnis R. E., Ewens W. J. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet. 1993 Mar;52(3):506–516. [PMC free article] [PubMed] [Google Scholar]
- Weeks D. E., Lange K. The affected-pedigree-member method of linkage analysis. Am J Hum Genet. 1988 Feb;42(2):315–326. [PMC free article] [PubMed] [Google Scholar]


