Abstract
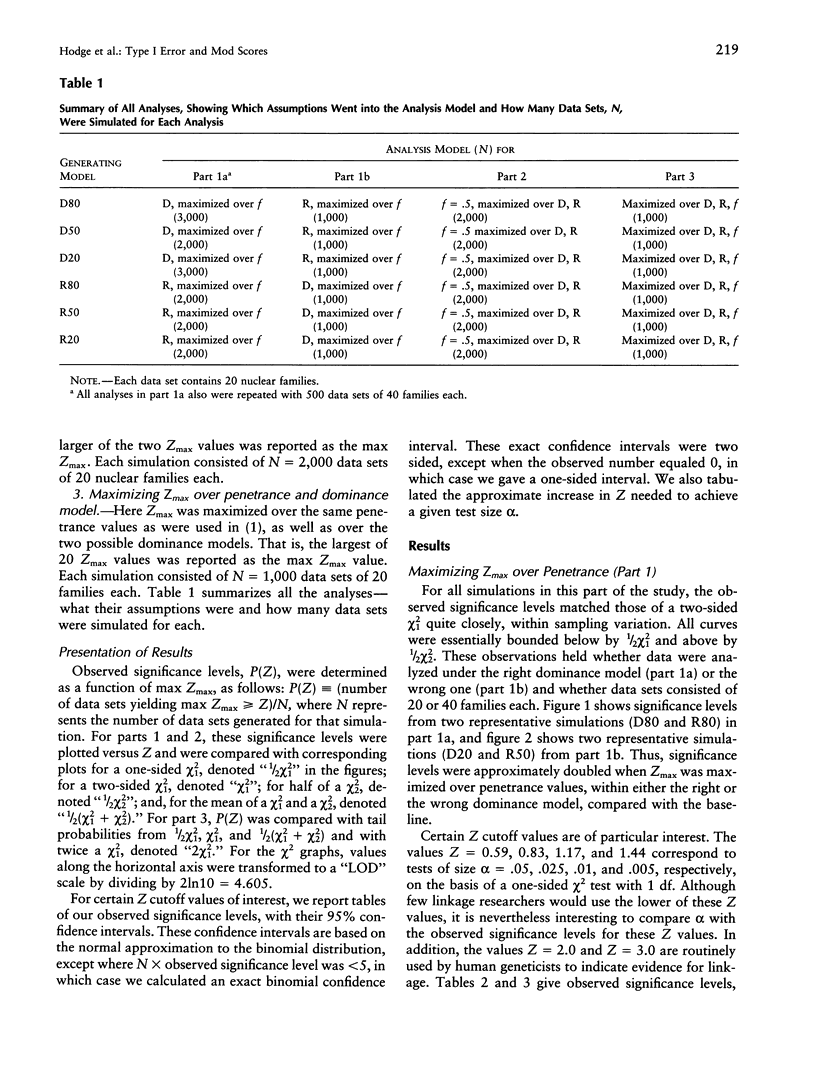
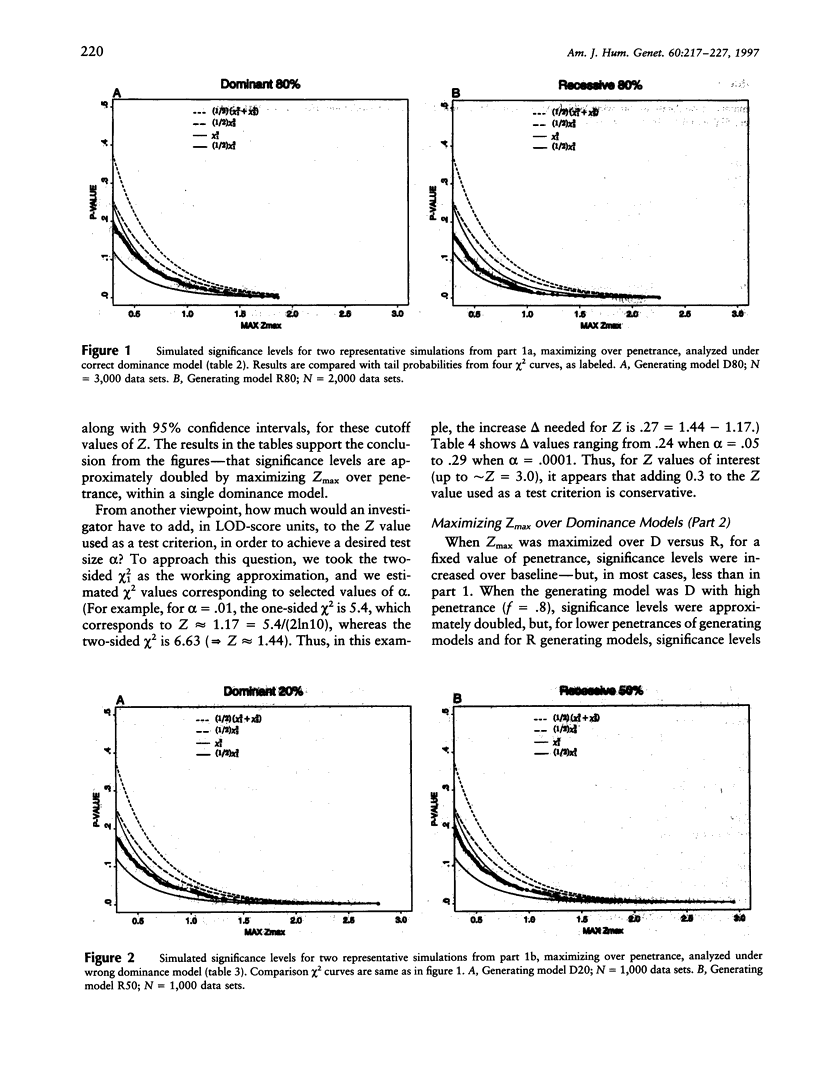
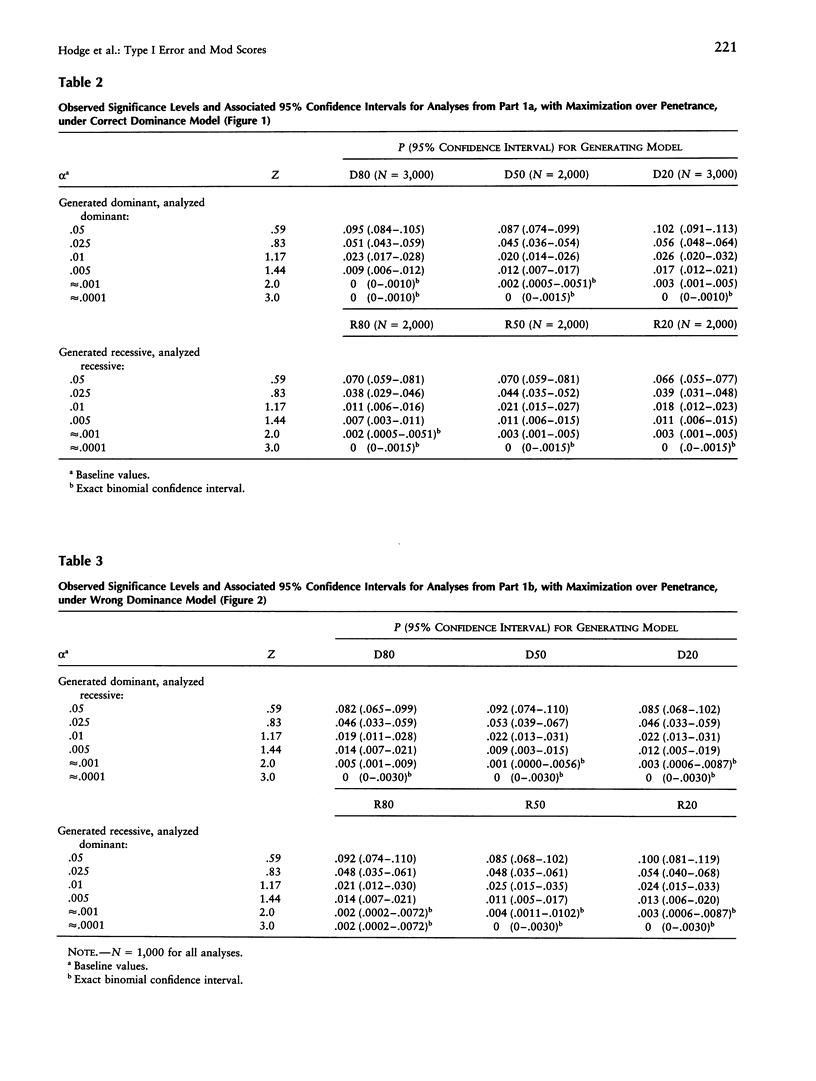
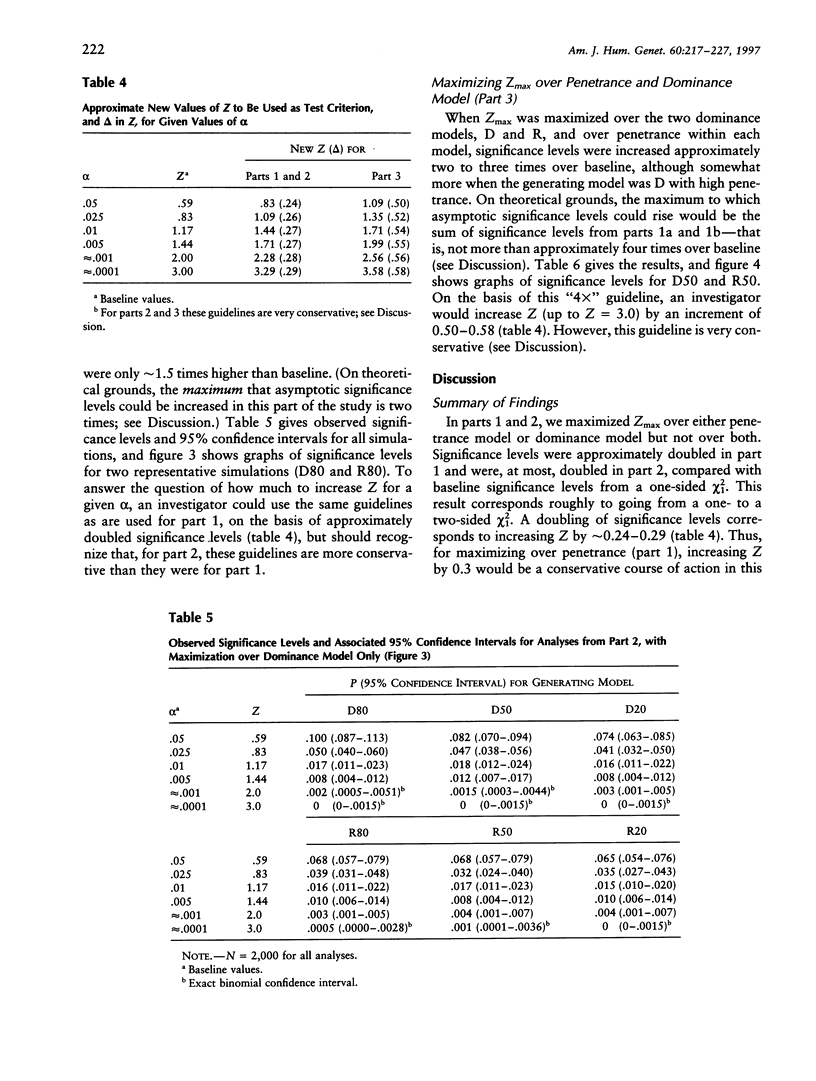
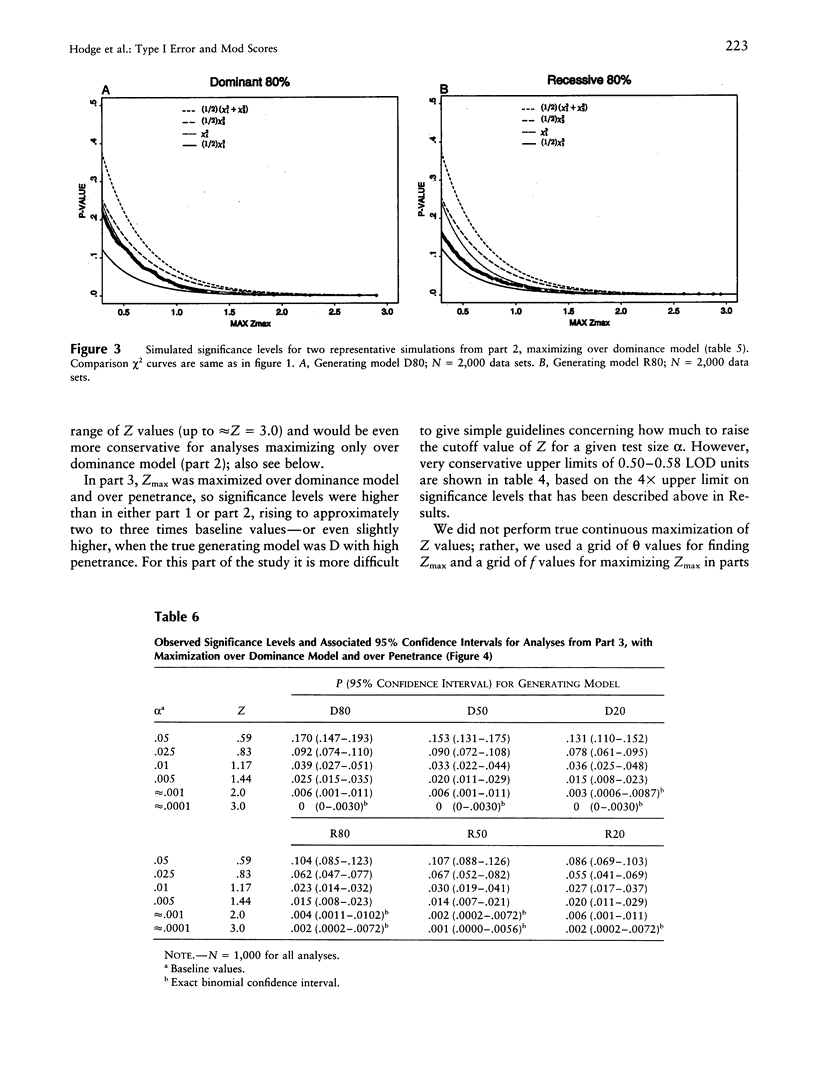
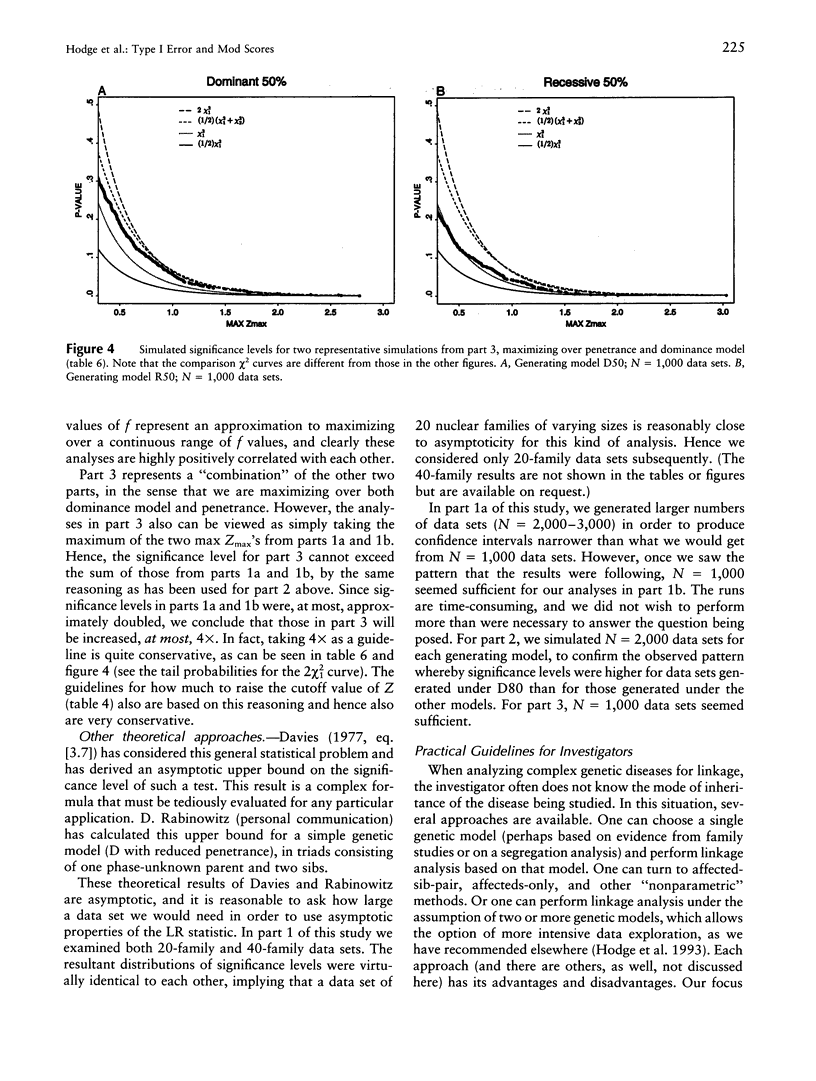
It is well known that maximizing the maximum LOD score over multiple parameter values or models (i.e., the method of mod scores, or MMLS), will inflate type I error, compared with assuming only one parameter value/model in the linkage analysis. On the other hand, a mod score often has greater power to detect linkage than does a LOD score (Z) calculated under a wrong genetic model. Therefore, it is of interest to determine the actual magnitude of type I error in realistic genetic situations. Simulated data sets with no linkage were generated under three dominant and three recessive single-locus models, with reduced penetrance (f = .8, .5, and .2). Data sets were analyzed for linkage by (1) maximizing over penetrance only, (2) maximizing over "dominance model" (i.e., dominant versus recessive), and (3) maximizing over both penetrance and dominance model simultaneously. In (1), the resultant significance levels were approximately doubled, compared with baseline values if one had not maximized over penetrances (i.e., compared with a one-sided chi2(1)). In (2), significance levels were increased somewhat less, and, in (3), they were increased by approximately two to three times (but not more than four times) over those of the one-sided chi2(1). This means that, for a given size of test alpha, an investigator would need to increase the Z used as a test criterion, by approximately 0.30 LOD units for analyses as in (1) or (2) and by 0.60 Z units for analyses as in (3). These guidelines, which are valid up to approximately Z = 3.0, are conservative for (1) and are very conservative for (2) and (3). By quantifying the increase in significance level (or, correspondingly, the increase in Z), our findings will enable users to rationally assess the advantages versus the disadvantages of mod scores.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clerget-Darpoux F., Babron M. C., Bonaïti-Pellié C. Assessing the effect of multiple linkage tests in complex diseases. Genet Epidemiol. 1990;7(4):245–253. doi: 10.1002/gepi.1370070403. [DOI] [PubMed] [Google Scholar]
- Clerget-Darpoux F., Bonaïti-Pellié C., Hochez J. Effects of misspecifying genetic parameters in lod score analysis. Biometrics. 1986 Jun;42(2):393–399. [PubMed] [Google Scholar]
- Clerget-Darpoux F., Bonaïti-Pellié C. Strategies based on marker information for the study of human diseases. Ann Hum Genet. 1992 May;56(Pt 2):145–153. doi: 10.1111/j.1469-1809.1992.tb01140.x. [DOI] [PubMed] [Google Scholar]
- Durner M., Greenberg D. A. Effect of heterogeneity and assumed mode of inheritance on lod scores. Am J Med Genet. 1992 Feb 1;42(3):271–275. doi: 10.1002/ajmg.1320420302. [DOI] [PubMed] [Google Scholar]
- Elston R. C. Man bites dog? The validity of maximizing lod scores to determine mode of inheritance. Am J Med Genet. 1989 Dec;34(4):487–488. doi: 10.1002/ajmg.1320340407. [DOI] [PubMed] [Google Scholar]
- Greenberg D. A., Berger B. Using lod-score differences to determine mode of inheritance: a simple, robust method even in the presence of heterogeneity and reduced penetrance. Am J Hum Genet. 1994 Oct;55(4):834–840. [PMC free article] [PubMed] [Google Scholar]
- Greenberg D. A., Hodge S. E. Linkage analysis under "random" and "genetic" reduced penetrance. Genet Epidemiol. 1989;6(1):259–264. doi: 10.1002/gepi.1370060145. [DOI] [PubMed] [Google Scholar]
- Greenberg D. A., Hodge S. E., Vieland V. J., Spence M. A. Affecteds-only linkage methods are not a panacea. Am J Hum Genet. 1996 Apr;58(4):892–895. [PMC free article] [PubMed] [Google Scholar]
- Greenberg D. A. Inferring mode of inheritance by comparison of lod scores. Am J Med Genet. 1989 Dec;34(4):480–486. doi: 10.1002/ajmg.1320340406. [DOI] [PubMed] [Google Scholar]
- Hodge S. E., Durner M., Vieland V. J., Greenberg D. A. Better data analysis through data exploration. Am J Hum Genet. 1993 Sep;53(3):775–777. [PMC free article] [PubMed] [Google Scholar]
- Hodge S. E., Elston R. C. Lods, wrods, and mods: the interpretation of lod scores calculated under different models. Genet Epidemiol. 1994;11(4):329–342. doi: 10.1002/gepi.1370110403. [DOI] [PubMed] [Google Scholar]
- Knapp M., Seuchter S. A., Baur M. P. Linkage analysis in nuclear families. 2: Relationship between affected sib-pair tests and lod score analysis. Hum Hered. 1994 Jan-Feb;44(1):44–51. doi: 10.1159/000154188. [DOI] [PubMed] [Google Scholar]
- MacLean C. J., Bishop D. T., Sherman S. L., Diehl S. R. Distribution of lod scores under uncertain mode of inheritance. Am J Hum Genet. 1993 Feb;52(2):354–361. [PMC free article] [PubMed] [Google Scholar]
- Ott J. Estimation of the recombination fraction in human pedigrees: efficient computation of the likelihood for human linkage studies. Am J Hum Genet. 1974 Sep;26(5):588–597. [PMC free article] [PubMed] [Google Scholar]
- Risch N. A note on multiple testing procedures in linkage analysis. Am J Hum Genet. 1991 Jun;48(6):1058–1064. [PMC free article] [PubMed] [Google Scholar]
- Sherrington R., Brynjolfsson J., Petursson H., Potter M., Dudleston K., Barraclough B., Wasmuth J., Dobbs M., Gurling H. Localization of a susceptibility locus for schizophrenia on chromosome 5. Nature. 1988 Nov 10;336(6195):164–167. doi: 10.1038/336164a0. [DOI] [PubMed] [Google Scholar]
- Vieland V. J., Greenberg D. A., Hodge S. E. Adequacy of single-locus approximations for linkage analyses of oligogenic traits: extension to multigenerational pedigree structures. Hum Hered. 1993 Nov-Dec;43(6):329–336. doi: 10.1159/000154155. [DOI] [PubMed] [Google Scholar]
- Vieland V. J., Hodge S. E., Greenberg D. A. Adequacy of single-locus approximations for linkage analyses of oligogenic traits. Genet Epidemiol. 1992;9(1):45–59. doi: 10.1002/gepi.1370090106. [DOI] [PubMed] [Google Scholar]
- Vieland V., Greenberg D. A., Hodge S. E., Ott J. Linkage analysis of two-locus diseases under single-locus and two-locus analysis models. Cytogenet Cell Genet. 1992;59(2-3):145–146. doi: 10.1159/000133229. [DOI] [PubMed] [Google Scholar]
- Weeks D. E., Lehner T., Squires-Wheeler E., Kaufmann C., Ott J. Measuring the inflation of the lod score due to its maximization over model parameter values in human linkage analysis. Genet Epidemiol. 1990;7(4):237–243. doi: 10.1002/gepi.1370070402. [DOI] [PubMed] [Google Scholar]
- Williamson J. A., Amos C. I. On the asymptotic behavior of the estimate of the recombination fraction under the null hypothesis of no linkage when the model is misspecified. Genet Epidemiol. 1990;7(5):309–318. doi: 10.1002/gepi.1370070502. [DOI] [PubMed] [Google Scholar]


