Summary
In eukaryotic cells, the turnover of long-lived proteins and large cytoplasmic structures is mediated by autophagy. Components that have to be eliminated are sequestered into double-membrane vesicles called autophagosomes and delivered into the lysosome or vacuole where they are destroyed by resident hydrolases. The integral membrane protein Atg9 is essential for both autophagy and the cytoplasm-to-vacuole targeting pathway, a selective biosynthetic process in Saccharomyces cerevisiae that is mechanistically and morphologically similar to autophagy. Atg9 cycles between the pre-autophagosomal structure, the putative site of double-membrane vesicle biogenesis and mitochondria. To understand the function of Atg9, and also its trafficking mode between these two locations, we identified mutants that affect specific Atg9 transport steps. We recently reported that five Atg proteins and phosphatidylinositol-3-phosphate regulate Atg9 recycling from the pre-autophagosomal structure. Here, we describe a different category of mutants that blocks Atg9 sorting from mitochondria. All mutants have been previously shown to be required for the normal progression of both the Cvt pathway and autophagy, but their precise role in these transport routes was unknown.
Keywords: Actin, Autophagy, Cytoplasm-to-vacuole targeting, Endoplasmic reticulum, Yeast
Introduction
The two principal pathways for protein catabolism in eukaryotic cells are proteasome-mediated degradation and autophagy. The proteasome is mostly used for the elimination of short-lived polypeptides, whereas autophagy is involved in the turnover of long-lived proteins. Another major difference between these two degradation systems is that the proteasome hydrolyzes single polypeptide chains one by one, whereas autophagy disposes of large protein aggregates, organelles and also entire bacteria in a single step by delivering its cargoes into the lysosome or vacuole interior where they are consumed by resident hydrolases (Levine and Klionsky, 2004; Reggiori and Klionsky, 2005). Because of this characteristic, autophagy is also able to degrade other cellular constituents such as lipids and carbohydrates.
The general mechanism of autophagy, and the most obvious morphological characteristic, is the sequestration of the cargoes destined to be eliminated into a cytosolic double-membrane vesicle called an autophagosome (Klionsky, 2004;Reggiori and Klionsky, 2005). Genetic screens in yeasts, in particular Saccharomyces cerevisiae, have led to the isolation of a large number of genes named ATG that are involved in autophagosome biogenesis (see Klionsky et al., 2003). Despite the fact that most of the Atg proteins are conserved among all eukaryotic organisms, some have only been identified at present in S. cerevisiae; accordingly, analyses in this system are necessary for the elucidation of their function (Reggiori and Klionsky, 2002; Reggiori and Klionsky, 2005). One of the advantages in using this model organism is that it possesses a biosynthetic and selective type of autophagy called the cytoplasm-to-vacuole targeting (Cvt) pathway that assures the delivery of two resident vacuolar hydrolases, aminopeptidase I (Ape1) and α-mannosidase (Hutchins and Klionsky, 2001; Klionsky et al., 1992). This transport route is mechanistically and morphologically similar to bulk autophagy and, therefore, it uses most of the same components, including Atg proteins (see Klionsky et al., 2003). In addition, other types of selective autophagy, including the specific degradation of peroxisomes (reviewed in Dunn et al., 2005), require almost all of the same Atg components as the Cvt pathway. Accordingly, the study of the Cvt pathway has provided important insights into the mechanism of autophagy.
Most of the Atg components appear to be primarily restricted to the pre-autophagosomal structure (PAS), the potential site of organization for autophagosome formation (Kim et al., 2002; Klionsky et al., 2003; Suzuki et al., 2001). However, despite the identification of the Atg proteins and the uncovering of several interactions among these factors, it remains unclear how autophagosomes are created. A major challenge in unveiling this process arises from the fact that the origin and the transport mode of the lipid bilayers employed to compose these structures are unknown (Reggiori, 2006). In an effort to solve this mystery, we have recently discovered that in the yeast S. cerevisiae, the transmembrane protein Atg9 cycles between the mitochondria and PAS (Reggiori et al., 2005b; Reggiori et al., 2004a). In addition, we have demonstrated that Atg9 retrieval transport from the PAS is regulated by the Atg1-Atg13 signaling complex and requires Atg2, Atg18 and the phosphatidylinisitol 3-phosphate [PtdIns(3)P] generated by the Atg14-containing PtdIns 3-kinase complex I (Reggiori et al., 2004a). In another yeast, Pichia pastoris, Atg9 displays a similar subcellular distribution and trafficking requirements, although some slight differences have been observed (Chang et al., 2005; Reggiori et al., 2005b). Here, we reveal that the dynamic Atg9 sorting from mitochondria is impaired when the morphology of this organelle is altered through mutations such as sec12 and vps52Δ that have previously been shown to block both the Cvt pathway and autophagy.
Results
Early secretion mutants disrupt Atg9 sorting from mitochondria
Retrieval of Atg9 from the PAS is regulated by the Atg1-Atg13 complex and requires Atg2, Atg18 and the PtdIns(3)P generated by the Atg14-containing PtdIns 3-kinase complex I (Reggiori et al., 2004a). The Atg9 sorting mechanism from mitochondria, however, remains uncharacterized. We reasoned that the mutation of certain genes could affect Atg9 exit from the mitochondria and its consequent delivery to the PAS. To gain further insight into the transport of Atg9 from the mitochondria we decided to examine mutant strains that have been shown to affect the Cvt pathway and autophagy.
The endoplasmic reticulum (ER) and mitochondria form reticular networks close to the plasma membrane and are closely apposed to one another (Prinz et al., 2000). The intimate association between these two organelles is also demonstrated by the presence of direct connections between them used for lipid transport and Ca2+ signaling (Daum and Vance, 1997; Rizzuto et al., 1998). Various studies have reported that yeast early secretion (sec) mutants have an indirect negative effect on both the Cvt pathway and autophagy (Hamasaki et al., 2003; Ishihara et al., 2001; Reggiori et al., 2004b). The ER structure is disorganized in these mutants, resulting in a severe disruption of the mitochondrial reticulum (Prinz et al., 2000). Because a morphological alteration of this organelle has an effect on Atg9 trafficking (Reggiori et al., 2005a), we decided to analyze the trafficking of this protein in the thermo sensitive strain sec12, which has previously been used to study both the Cvt pathway and autophagy (Hamasaki et al., 2003; Ishihara et al., 2001; Reggiori et al., 2004b). The SEC12 gene encodes a GDP-GTP exchange factor that it is required for vesicle budding and exit from the ER (Barlowe and Schekman, 1993).
We first verified that we could reproduce and monitor the same conditions that led to the disruption of the mitochondrial network. To do so, wild-type and sec12 strains carrying a plasmid that expressed the ER protein marker Spo7-GFP were grown at a permissive temperature (24°C) to early logarithmic (log)-phase before being transferred to a nonpermissive temperature (37°C) for 2 hours. Cells were also incubated with MitoFluor Red, a dye that permits visualization of mitochondria (Reggiori et al., 2005b). At each time point a stack of images with focal planes 0.15 μm apart was obtained with a wide-field optical sectioning microscope and deconvolved; the focal plane was either close to the center or the periphery of the cell. As expected, the reticular conformation of the cortical ER could be seen in the wild-type strain grown at both permissive and restrictive temperatures (Fig. 1). The usual tubular arrangement of the mitochondria is also evident in the same cells. The ER and mitochondrial morphology in the sec12 mutant was indistinguishable from that of the wild type when cells were kept at 24°C (Fig. 1). As reported, the transfer of these cells to 37°C for 2 hours provoked the dissolution of the cortical ER network. The GFP signal at the periphery of the cells became more homogeneously distributed. One of the consequences associated with this alteration was the fragmentation of the mitochondrial tubular network (Fig. 1) (Prinz et al., 2000).
Fig. 1.
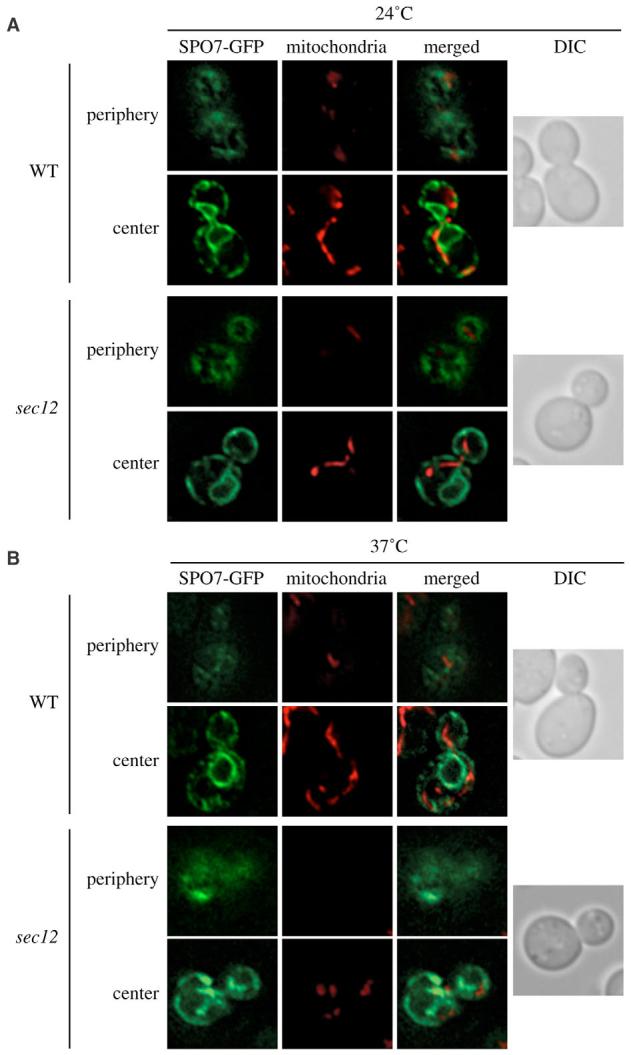
Early sec mutants block cycling of Atg9. The structure of the peripheral ER and the mitochondrial reticulum is altered in the sec12 mutant at nonpermissive temperatures. (A,B) The wild-type (SEY6210) and sec12 (FBY217) strains both transformed with the plasmid expressing Spo7-GFP (YEplac122-TRP1-SPO7-GFP) were grown at 24°C (A) and then transferred to 37°C (B) for 2 hours. Cells were labeled with MitoFluor Red and viewed before and after the temperature shift. Images of the fluorescent signal were acquired while focusing on either the center or the periphery of the cells. DIC, differential-interference-contrast images.
We then analyzed whether the effects of the sec12 mutation on mitochondrial morphology affected Atg9 sorting from this organelle. This protein is entirely accumulated at the PAS in the absence of genes such as ATG1, which are essential for its retrograde transport from this structure (Reggiori et al., 2004a). Condensation of fluorescent chimeric Atg9 proteins into a single bright dot can be used to easily detect this trafficking defect. It has been recently shown that the accumulation of Atg9 in the atg1Δ strain can be prevented by the deletion of genes epistatic to ATG1 and required for Atg9 delivery to the PAS by using the transport of Atg9 after knocking-out Atg1 (TAKA) assay (Cheong et al., 2005; Reggiori et al., 2005a; Shintani and Klionsky, 2004). In these double mutants, fluorescent Atg9 remains disseminated in several punctate structures. To investigate whether the sec12 allele blocked Atg9 transport from the mitochondria to the PAS, we generated a sec12 atg1Δ double-mutant strain expressing Atg9-YFP and carrying a plasmid encoding the atg1ts allele; a simultaneous inactivation of Sec12 and Atg1 is established when this strain is incubated at 37°C. Controls were sec12 and atg1ts single mutants that both expressed Atg9-YFP. All cells were grown at 24°C to an early log-phase and then transferred to 37°C for 1 hour.
When all three strains were grown at 24°C, Atg9-YFP was dispersed in several punctate structures, some of which colocalized with mitochondria (Fig. 2A). As expected, Atg9-YFP accumulated as a single punctate dot at the PAS in the atg1ts strain after the temperature shift (Fig. 2B). The same temperature change caused the dispersal of Atg9-YFP clusters on mitochondria in sec12 cells, leading to the localization of this chimera all along the outer membrane of this organelle. An identical result was obtained in the atg1ts sec12 double mutant revealing that the sec12 mutation is epistatic to atg1ts. To generalize this observation to all the early sec-type mutants, we repeated the same experiment in another of these mutants, sed5. The SED5 gene encodes a tSNARE that it is required for the fusion of ER-derived vesicles with the Golgi complex (Hardwick and Pelham, 1992). As shown in Fig. 2C, Atg9 trafficking is also blocked in the atg1ts sed5 double mutant.
Fig. 2.
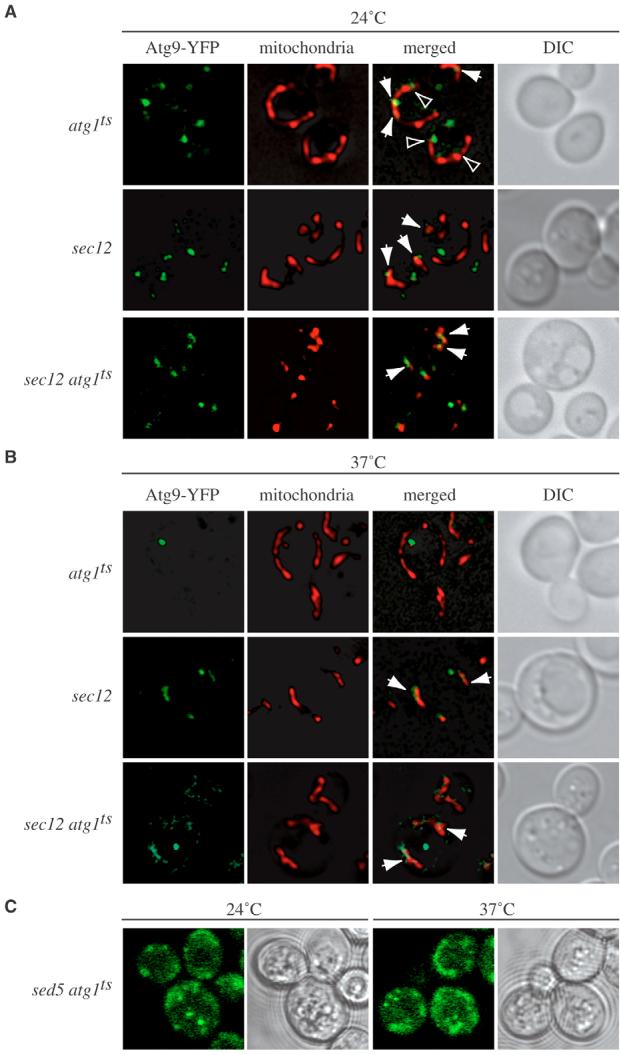
Atg9 sorting from mitochondria is impaired in sec12 and sed5 cells. (A,B,C) The sec12 atg1Δ Atg9-YFP (FRY237) or sed5 atg1Δ (FRY303) strains transformed with the vector carrying the atg1ts allele were grown at permissive temperature (24°C) (A; C, left panels) and then shifted to 37°C (B; C, right panels) for 1 hour. Cells were photographed before and after the temperature change; the cells in C were first subjected to mild fixation. (A,B) To visualize mitochondria, MitoFluor Red was added to the culture medium 30 minutes before taking the pictures. The YFP signal is green to facilitate comparison with MitoFluor Red. Control strains were atg1Δ Atg9-YFP (FRY138) carrying the plasmid expressing Atg1ts and sec12 Atg9-YFP (FRY154). Arrows indicate colocalization of Atg9-YFP and MitoFluor Red. Arrowheads indicate regions where Atg9-YFP is in close proximity to MitoFluor Red. DIC, differential-interference-contrast images.
Actin filaments are essential to maintain the mitochondrial reticulum morphology and for its correct segregation during cell division (Boldogh et al., 1998; Drubin et al., 1993). Consequently, their dissolution severely affects the organization of this organelle and one of the consequences is an impairment of Atg9 trafficking (Reggiori et al., 2005a). We decided to investigate whether the disruption of the mitochondrial reticulum in the sec12 mutant incubated at restrictive temperature was caused by a defect in the formation of actin cables. The sec12 strain was grown to an early log phase at 24°C or shifted to 37°C for 90 minutes, fixed and stained with the Texas Red-phalloidin fluorescent conjugate to visualize actin cables. As a control, the wild-type strain was grown at 24°C and treated in the same way. As expected, the wild-type strain showed the presence of both actin cables aligned with the cell division axis and actin patches (Fig. 3). Similar fluorescence images were obtained in sec12 mutants incubated at both permissive and restrictive temperatures. We concluded that in this mutant, the disruption of the mitochondrial network in not due to the dissolution or incorrect alignment of actin cables.
Fig. 3.
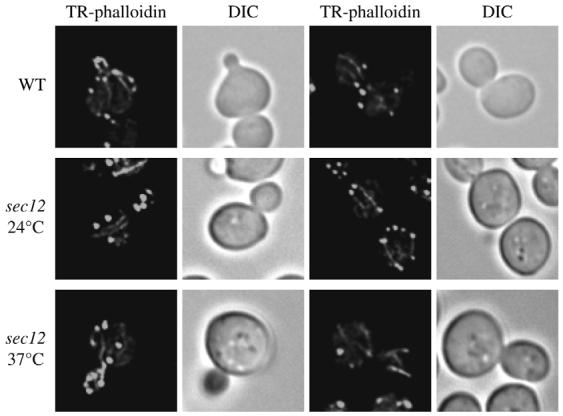
Organization of actin cables is not affected in sec12 cells. The wild-type (SEY6210) and sec12 (FBY217) strains grown at 24°C were fixed, permeabilized and incubated with Texas Red-phalloidin (TR-phalloidin) as described in Materials and Methods before collecting fluorescence images. The sec12 mutant was also treated and imaged as above after being incubated at restrictive temperature for 90 minutes. DIC, differential-interference-contrast images.
Atg9 trafficking is impaired in mutants that lack the VFT complex in growing conditions
The Cvt pathway is completely blocked and autophagy is reduced in the absence of any of the Vps fifty-three (VFT) tethering-complex subunits (Reggiori et al., 2003). This set of factors is also essential for efficient protein passage through the Golgi complex because it catalyzes retrieval transport to the late compartments of this organelle and therefore guarantees proper functioning (Conibear and Stevens, 2000). The contribution of the VFT complex in double-membrane vesicle formation is not clear, but because both the Golgi complex and the ER play an important mutual role in maintaining their correct morphologies, we decided to explore whether VFT-complex deletion mutants had an Atg9 trafficking defect similar to that observed in the sec12 mutant.
We first investigated ER and mitochondria morphology in the absence of Vps52, one of the four VFT-complex subunits (Conibear and Stevens, 2000; Reggiori et al., 2003). Wild-type and vps52Δ strains carrying the plasmid expressing Spo7-GFP were grown to an early log-phase in rich medium, stained with MitoFluor Red and imaged (see Fig. 1). Wild-type and vps52Δ cells had an almost identical ER structure, a tighter cortical network in the mutant being the only difference (Fig. 4). Mitochondrial tubules were completely fragmented and often clumped together in the absence of VPS52 (Fig. 4, Fig. 5A). Because autophagy is only partially affected in mutants that lack the VFT complex, the same microscopy analysis was repeated after depriving the cells of nitrogen for 2 hours. Starvation did not provoke major morphological changes of the ER in wild-type or mutant strains (Fig. 4), and most of the mitochondrial network was restored in vps52Δ cells under these conditions.
Fig. 4.
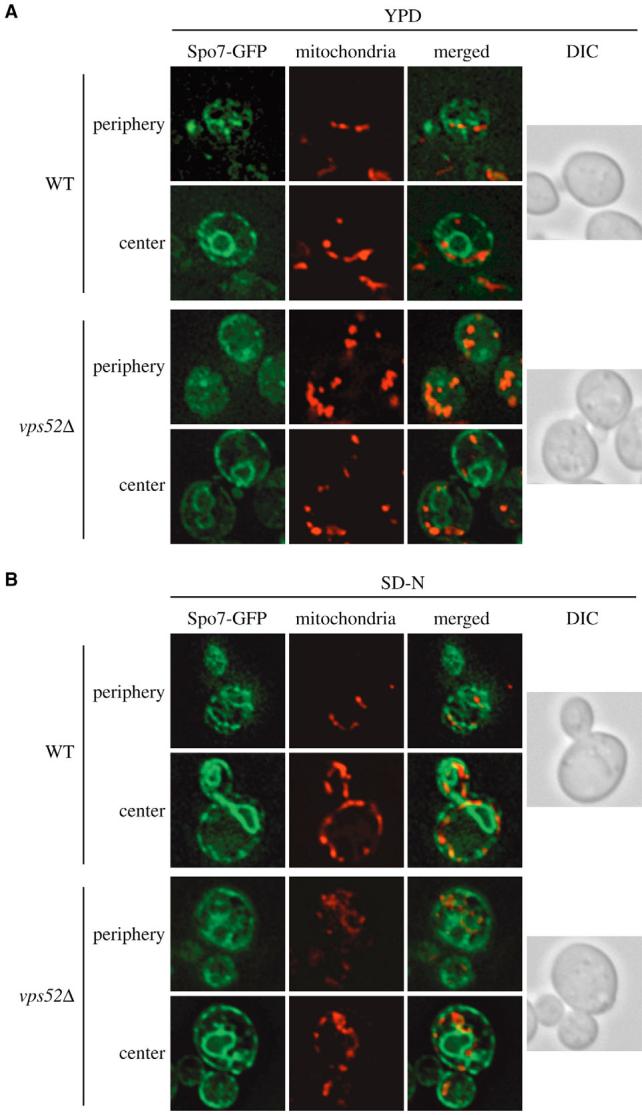
Atg9 sorting from mitochondria is impaired in the absence of the VFT complex. The wild-type (SEY6210) and vps52Δ (PSY113) strains both transformed with the plasmid expressing Spo7-GFP (YEplac122-TRP1-SPO7-GFP) were grown at 30°C to an early log-phase in rich medium (A) or starved of nitrogen for 2 hours (B). Cells were labeled with MitoFluor Red and imaged as described in Fig. 1. The mitochondrial reticulum was disrupted in the absence of VPS52 in rich medium. DIC, differential-interference-contrast images.
Fig. 5.
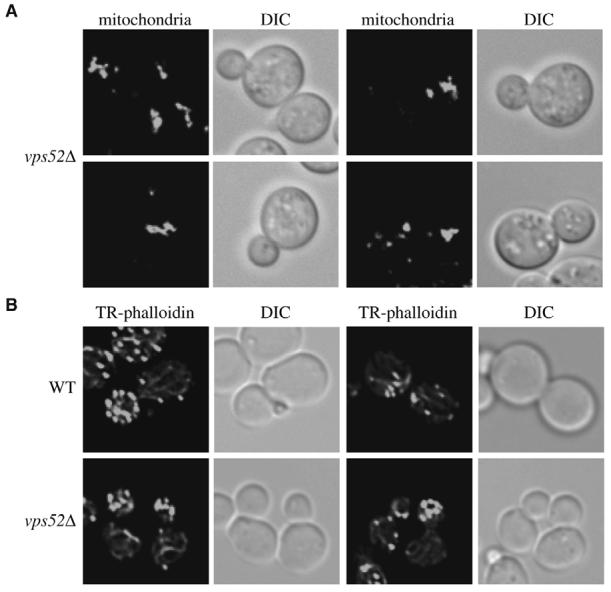
Alteration of subcellular structures in the vps52Δ mutant. (A) Mitochondrial morphology is severely compromised in the absence of Vps52. The vps52Δ (PSY113) strain was grown in rich medium to an early log-phase and mitochondria labeled by adding MitoFluor Red prior to imaging. (B) Actin cables are aberrantly organized in the vps52Δ mutant. Cells grown as in panel A were fixed, permeabilized and incubated with Texas Red-phalloidin (TR-phalloidin) as described in Materials and Methods before collecting fluorescence images. DIC, differential-interference-contrast images.
It was recently reported that, in the absence of Vps54-another VFT-complex subunit (Conibear and Stevens, 2000; Reggiori et al., 2003)-actin cables are aberrantly arranged, provoking defects in several actin-related processes (Fiedler et al., 2002). Consequently, we decided to verify the subcellular positioning of these structures in the vps52Δ strain because the incorrect organization of actin could explain why the mitochondrial reticulum was altered. Wild-type and vps52Δ cells were grown to an early log-phase, fixed and stained with the Texas Red-phalloidin fluorescent conjugate to visualize actin cables. Once more, the wild-type strain showed the presence of actin cables aligned with the cell division axis (Fig. 5B). The same structures were also present in the vps52Δ mutant, but their positioning was completely random. We concluded that in the absence of Vps52 and, by extension, Vps51, Vps53 and Vps54, actin cables are aberrantly organized, which compromises the mitochondrial reticulum morphology.
Having already determined that the proper mitochondrial organization is essential for Atg9 trafficking (Fig. 2) (Reggiori et al., 2005a), we decided to examine whether the altered morphology of this organelle in the vps52Δ strain had a similar effect. To do that, we again took advantage of the TAKA assay. An atg1Δ vps52Δ double-mutant strain expressing ATG9-YFP and the corresponding single deletion mutants were imaged after being either grown in rich medium or nitrogen-starved for 3 hours. As expected, Atg9-YFP was concentrated at the PAS in the absence of ATG1 and this localization pattern was unaffected by nitrogen deprivation (our unpublished observations) (Reggiori et al., 2004a). By contrast, and similar to wild-type cells (data not shown), this chimera was distributed to several punctate structures in the vps52Δ strain in both rich and starvation media (Fig. 6). The ATG1 deletion in the vps52Δ background had no effect on Atg9-YFP localization when cells were kept in rich medium, revealing that the vps52Δ defect was epistatic to the atg1Δ mutation. Atg9-YFP accumulated at the PAS when the double deletion mutant was transferred for at least 3 hours to medium lacking nitrogen, indicating that starvation conditions bypassed the Atg9 cycling defect of the vps52Δ strain (Fig. 6). This observation is consistent with the fact that the VFT complex plays a marginal role during autophagy (Reggiori et al., 2003).
Fig. 6.
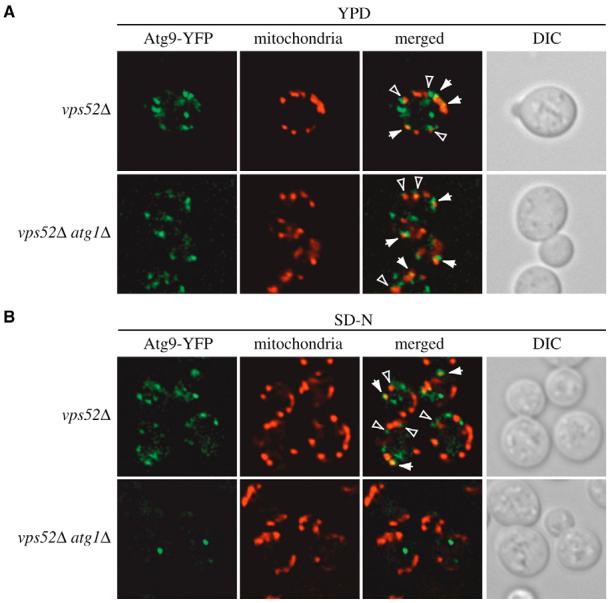
Atg9 sorting from mitochondria is blocked in the vps52Δ strain in rich medium but not under starvation conditions. (A,B) The vps52Δ Atg9-YFP (FRY238) and vps52Δ atg1Δ Atg9-YFP (FRY192) strains were grown in rich medium (A) or starved in SD-N for 3 hours (B) and then photographed. Mitochondria were labeled by adding MitoFluor Red 30 minutes prior to imaging. Arrows indicate sites of colocalization between Atg9-YFP and MitoFluor Red. Arrowheads indicate regions where Atg9-YFP is in close proximity to MitoFluor Red. DIC, differential-interference-contrast images.
As a control, to demonstrate that the Atg9 trafficking impairment observed in the vps52Δ strain is not a common characteristic of all mutants with a vacuolar protein sorting (vps) defect, we analyzed Atg9 transport in vps4Δ cells. Vps4 is an AAA-type ATPase essential for multivesicular body formation and, in its absence, abnormally enlarged late endosomes accumulate that can be visualized with the endocytic dye FM 4-64 (Babst et al., 1997). Accordingly, vps4Δ and atg1Δ vps4Δ mutants expressing ATG9-YFP were fixed and then imaged after being grown in rich medium in the presence of fixable FM 4-64 (FM 4-64 FX) for 1 hour. As shown in Fig. 7, the Atg9-YFP distribution in the vps4Δ deletion mutant was similar to that in wild-type cells, whereas the same chimera was concentrated at the PAS in the absence of ATG1. Thus, we concluded that Vps4 is not involved in Atg9 trafficking. This result is in complete agreement with previous data that have shown that Vps4 is not required for the Cvt pathway and demonstrates the specificity of the vps52Δ mutation (Epple et al., 2003; Reggiori et al., 2004b).
Fig. 7.
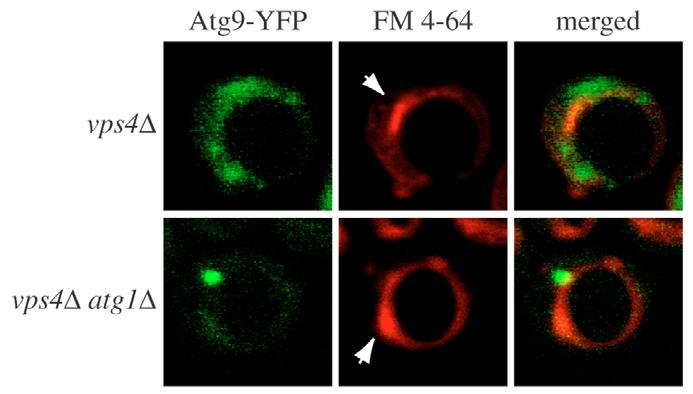
Atg9 trafficking from mitochondria is not affected in the absence of Vps4. The vps4Δ Atg9-YFP (FRY304) and vps4Δ atg1Δ Atg9-YFP (FRY305) strains were grown in rich medium, subjected to mild fixation and then photographed. Vacuoles were labeled by adding FM 4-64 FX 1 hour prior to imaging. The YFP signal is in green to facilitate comparison with FM 4-64 FX. Arrows indicate abnormally enlarged late endosomes typical of the vps4Δ mutant.
Discussion
The integral membrane protein Atg9 cycles between the PAS and mitochondria and is essential for both the Cvt pathway and autophagy (Noda et al., 2000; Reggiori et al., 2005b; Reggiori et al., 2004a). To understand the function of this factor and also its trafficking mode between these two locations, we decided to identify mutants that affect specific Atg9 transport steps. We have recently reported that five Atg proteins and PtdIns(3)P regulate Atg9 recycling from the PAS (Fig. 8A) (Reggiori et al., 2004a). Here, we described a different category of mutants that blocked Atg9 sorting from mitochondria and, consequently, its delivery to the PAS (Fig. 8). Interestingly, the early sec and VFT mutants have previously been shown to affect both the Cvt pathway and autophagy but their effective role in these transport routes was unknown because they do not play a direct role in double-membrane vesicle formation (Hamasaki et al., 2003; Ishihara et al., 2001; Reggiori et al., 2004b; Reggiori et al., 2003). In our study we analyzed representatives of each of these two groups, sec12, sed5 and vps52Δ, and show that they block Atg9 sorting from mitochondria by compromising the normal morphology of this organelle (Figs 1, 2, 4, 5, Fig. 3A). Our data, however, cannot eliminate the possibility that other Cvt pathway and autophagy steps are also impaired in these mutants.
Fig. 8.
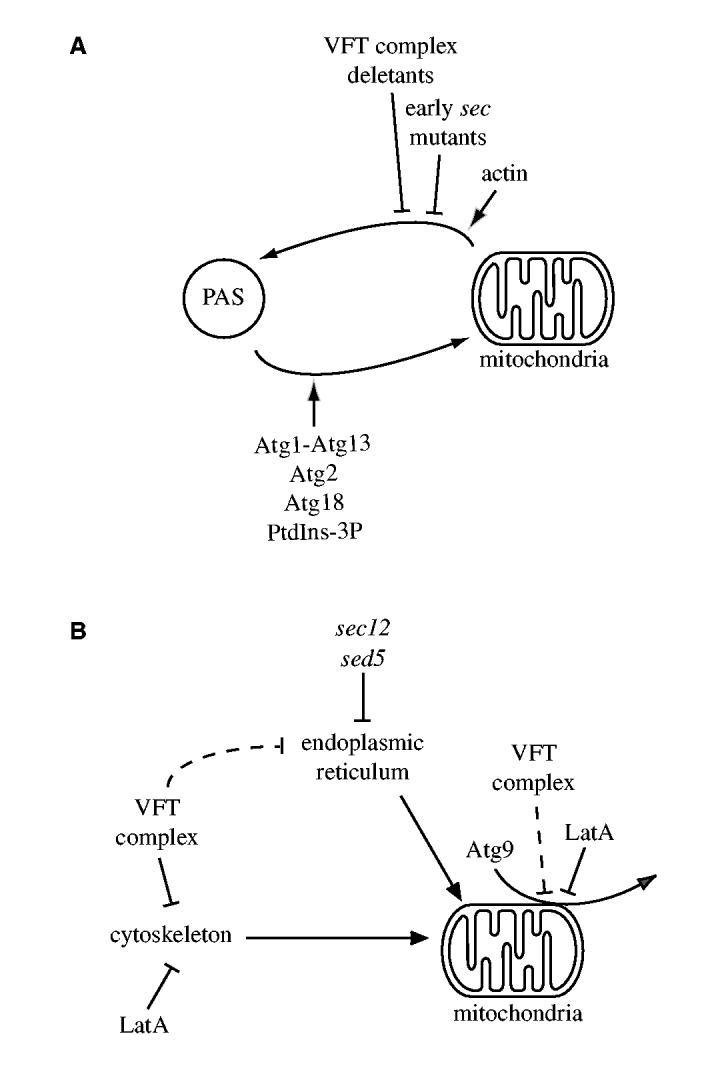
Model of subcellular Atg9 trafficking in S. cerevisiae. (A) Atg9 cycles between the PAS and mitochondria. Atg9 sorting from mitochondria is blocked by disruption of the ER-mitochondrial network by early sec mutations, deletion mutants that lack VFT-complex subunit and actin filament disruption. Once the double-membrane vesicle is completed and the function of Atg9 is no longer needed, the Atg1-Atg13 complex triggers the retrieval transport of this protein from the PAS (Reggiori et al., 2004a). Atg2, Atg18 and the PtdIns(3)P generated by the Atg14-containing PtdIns 3-kinase complex I also participate in this recycling event. (B) Inhibition of the Atg9 sorting event from mitochondria. The proper organization of the mitochondrial reticulum is maintained by both the cytoskeleton and a direct connection with the ER. Alteration of one of these structures provokes the disruption of the mitochondrial network, which blocks Atg9 trafficking. LatA treatment and deletion of each one of the subunits of the VFT complex affects actin filaments, whereas early sec mutations disrupt the ER peripheral network. In addition, actin also plays a direct role in Atg9 delivery and cargo recruitment to the PAS (Reggiori et al., 2005a).
In the absence of VFT-complex subunits, the mitochondrial network is dramatically compromised but the ER morphology is not seriously altered (Fig. 4, Fig. 5A). It is not known why, but actin cables are aberrantly arranged in the absence of VFT-complex subunits (Fig. 5B) (Fiedler et al., 2002), and actin plays a direct role in maintaining the normal organization of the mitochondrial reticulum (Drubin et al., 1993). Mutations in this polymeric protein create clumps of mitochondria that resemble those observed in vps52Δ cells (Fig. 4, Fig. 5A) (Drubin et al., 1993), and one of the consequences is the failure of Atg9 to reach the PAS (Fig. 4). An identical result was obtained when yeast cells were incubated with latrunculin A (LatA) (Reggiori et al., 2005a). That result supports the conclusion from the current study, which shows the importance of actin in maintaining mitochondrial morphology, thus allowing proper Atg9 cycling and progression of the Cvt pathway. LatA is a toxin that inhibits actin polymerization and, therefore, disrupts the cytoskeleton and mitochondrial morphology (Ayscough et al., 1997; Boldogh et al., 1998; Spector et al., 1983). However, actin seems to be also involved in the cargo-recruitment step of the Cvt pathway (Reggiori et al., 2005a). Consequently, we cannot exclude that this step is also affected in the vps52Δ mutant. It has previously been shown that the prApe1 transport block is reversed when the vps52Δ strain is starved for nitrogen (Reggiori et al., 2003). The mitochondrial reticulum seemed largely reestablished and, consequently, Atg9 trafficking was also restored under the same conditions, explaining the previous observation (Fig. 6).
In early sec mutants, the ER expansion that is accompanied by the alteration of several of its functions provokes the disruption of the mitochondrial reticulum (Fig. 1) (Prinz et al., 2000). In agreement with the results obtained with the vps52Δ deletion mutant and LatA treatment (Fig. 6) (Reggiori et al., 2005a), the change of the mitochondrial morphology causes an Atg9 transport defect (Fig. 2). In the sec12 mutant, however, Atg9 became homogeneously distributed along the mitochondrial outer membrane, whereas the same factor remained assembled in discrete punctate structures scattered on the surface of this compartment in the vps52Δ strain (Figs 1, 2, 4, 6). The reason for this discrepancy is unclear, but one possibility is that the two mutations have different effects on mitochondrial physiology. Because mitochondria are structurally and functionally connected with the ER (Daum and Vance, 1997; Prinz et al., 2000; Rizzuto et al., 1998), the sec12 allele probably impairs several of the functions of this organelle. As mentioned, actin has at least two functions in the Cvt pathway: Maintaining the mitochondrial structure and recruitment of the cargo (Reggiori et al., 2005a). Actin filaments and cables were present in sec12 cells incubated at restrictive temperature (Fig. 3), which is why this mutant enabled us to show that normal functionality of the mitochondrial reticulum is a requirement for the Atg9 sorting from this organelle and, therefore, double-membrane vesicle biogenesis.
The impairment of Atg9 trafficking in sec12 or sed5 cells and VFT-complex mutants probably causes the formation of an aberrant or not completed PAS. It is interesting to note that, the Cvt complex is not correctly recruited to the PAS in both sec12 and vps52Δ strains, supporting this idea and confirming that both are part of the same class of mutants (Reggiori et al., 2004b; Reggiori et al., 2003). A similar observation has also been made in LatA-treated cells (Reggiori et al., 2005a).
In conclusion, our study has revealed which step of double-membrane vesicle formation is affected by a set of mutants previously shown to block this process, by showing that they impair sorting of Atg9 from mitochondria (Fig. 8). These observations provide the opportunity to specifically block Atg9 trafficking at a specific stage and allow us to design future experiments that will facilitate our understanding of the Atg9 sorting mechanism from mitochondria, an important part of the membrane dynamic during autophagy.
Materials and Methods
Strains, media and plasmids
The S. cerevisiae strains used in this study are listed in Table 1. For gene disruptions, the entire ATG1, VPS52 and VPS4 coding regions were replaced with either the Escherichia coli kanr, S. cerevisiae TRP1, Kluyveromyces lactis URA3 or LEU2 genes using PCR primers containing ∼40 bases of identity to the regions flanking the open reading frame. PCR-based integration of YFP at the 3′ end of ATG9 was used to generate strains expressing fusion proteins under the control of their native promoters. The template for integration was pDH3 (Drees et al., 2001). PCR verification and prApe1 processing were used to confirm the functionality of all genomic fusions. Plasmids expressing Spo7-GFP (YEplac122-TRP1-SPO7-GFP) and Atg1ts have been described elsewhere (Siniossoglou et al., 1998; Suzuki et al., 2001).
Table 1.
Strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| FBY217 | MATa sec12-4 his3 leu2 ura3 trp1 ade2 | A. Conzelmann |
| FRY138 | SEY6210 ATG9-YFP::HIS3 S.k. atg1Δ::URA3 | (Reggiori et al., 2004a) |
| FRY154 | MATa sec12-4 his3 leu2 ura3 trp1 ade2 ATG9-YFP::HIS3 S.k. | This study |
| FRY162 | SEY6210 ATG9-GFP::HIS3 S.k. | (Reggiori et al., 2005b) |
| FRY192 | SEY6210 ATG9-YFP::HIS3 S.k. vps52Δ::TRP1 atg1Δ::URA3 | This study |
| FRY237 | MATa sec12-4 his3 leu2 ura3 trp1 ade2 ATG9-YFP::HIS3 S.k. atg1Δ::URA3 | This study |
| FRY238 | SEY6210 ATG9-YFP::HIS3 S.k. vps52Δ::URA3 K.l. | This study |
| FRY239 | SEY6210 ATG9-YFP::KAN atg1Δ::HIS3 S.k. | This study |
| FRY303 | SEY6210 sed5-1 ATG9-YFP::HIS3 S.k. atg1Δ::URA3 | This study |
| FRY304 | SEY6210 ATG9-YFP::HIS3 S.k. vps4Δ::LEU2 K.l. | This study |
| FRY305 | SEY6210 ATG9-YFP::HIS3 S.k. atg1Δ::HIS3 S.k. vps4Δ::LEU2 K.l. | This study |
| PSY113 | SEY6210 vps52Δ::HIS3 S.k. | (Reggiori et al., 2003) |
| SEY6210 | MAT αura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel GAL | (Robinson et al., 1988) |
Yeast cells were grown in rich medium (YPD; 1% yeast extract, 2% peptone, 2% glucose) or synthetic minimal medium (SMD; 0.67% yeast nitrogen base, 2% glucose, amino acids and vitamins as needed). Starvation experiments were conducted in synthetic medium lacking nitrogen (SD-N; 0.17% yeast nitrogen base without amino acids, 2% glucose).
Fluorescence microscopy
Yeast cells were grown or starved in the appropriate medium before imaging. Fluorescent labeling of mitochondria with MitoFluor Red 589 (Molecular Probes, Eugene, OR) and actin cytoskeleton staining with Texas Red®-X phalloidin (Texas Red-phalloidin, Molecular Probes) were conducted as described previously (Reggiori et al., 2005a; Reggiori et al., 2005b). Fluorescence signals were visualized with a DeltaVision Spectris fluorescence microscope (Applied Precision, Issaquah, WA). The images were captured with a CoolSnap camera and deconvolved by using SoftWoRx software (Applied Precision).
The images of cells subjected to mild fixation (Reggiori et al., 2005b) shown in Fig. 2C and Fig. 7 were captured with a confocal scanning microscope (Leica TCS-NT, Heidelberg, Germany). For visualization of the vacuole, 10 μM of fixable FM 4-64 (FM 4-64 FX, Molecular Probes) was added 1 hour prior to fixation.
Acknowledgments
We thank Andreas Conzelmann, Yoshinori Ohsumi, Hugh Pelham and Symeon Siniossoglou for reagents. This work was supported by the National Institutes of Health Public Health Service grant GM53396 (to D.J.K.).
References
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Boldogh I, Vojtov N, Karmon S, Pon LA. Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J. Cell Biol. 1998;141:1371–1381. doi: 10.1083/jcb.141.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T, Schroder LA, Thomson JM, Klocman AS, Tomasini AJ, Stromhaug PE, Dunn WA., Jr PpATG9 encodes a novel membrane protein that traffics to vacuolar membranes, which sequester peroxisomes during pexophagy in Pichia pastoris. Mol. Biol. Cell. 2005;16:4941–4953. doi: 10.1091/mbc.E05-02-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang C-W, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol. Biol. Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Vance JE. Import of lipids into mitochondria. Prog. Lipid Res. 1997;36:103–130. doi: 10.1016/s0163-7827(97)00006-4. [DOI] [PubMed] [Google Scholar]
- Drees BL, Sundin B, Brazeau E, Caviston JP, Chen GC, Guo W, Kozminski KG, Lau MW, Moskow JJ, Tong A, et al. A protein interaction map for cell polarity development. J. Cell Biol. 2001;154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Jones HD, Wertman KF. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol. Biol. Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WA, Jr, Cregg JM, Kiel JAKW, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Epple UD, Eskelinen E-L, Thumm M. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 2003;278:7810–7821. doi: 10.1074/jbc.M209309200. [DOI] [PubMed] [Google Scholar]
- Fiedler TA, Karpova TS, Fleig U, Young ME, Cooper JA, Hegemann JH. The vesicular transport protein Cgp1p/Vps54p/Tcs3p/Luv1p is required for the integrity of the actin cytoskeleton. Mol. Genet. Genom. 2002;268:190–205. doi: 10.1007/s00438-002-0748-4. [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Noda T, Ohsumi Y. The early secretory pathway contributes to autophagy in yeast. Cell Struct. Funct. 2003;28:49–54. doi: 10.1247/csf.28.49. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HRB. SED5 encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J. Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang W-P, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy. Landes Bioscience; Georgetown, TX: 2004. [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F. The membrane origin for autophagy. Curr. Top. Dev. Biol. 2006;74:1–30. doi: 10.1016/S0070-2153(06)74001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryotic Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagosomes: Biogenesis from scratch? Curr. Opin. Cell Biol. 2005;17:415–422. doi: 10.1016/j.ceb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Wang C-W, Stromhaug PE, Shintani T, Klionsky DJ. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 2003;278:5009–5020. doi: 10.1074/jbc.M210436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell. 2004a;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Wang C-W, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004b;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005a;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005b;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 1998;17:6449–6464. doi: 10.1093/emboj/17.22.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I, Shochet NR, Kashman Y, Groweiss A. Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]


