Summary
In eukaryotes, distinct regions of the genome are packaged as euchromatin (less condensed, more active) or heterochromatin (condensed, silenced). Studies in yeast, plants and flies suggest that RNA interference (RNAi) is linked to heterochromatin formation and transcriptional silencing of transposable element (TE) sequences [1, 2]. We previously reported that insertion of a mobile hsp70-white reporter within 10 kb of a 1360 element on chromosome four of Drosophila melanogaster correlates with variegation (silencing) [3]. Here we report small RNAs (∼23 nt) corresponding to 1360, indicating processing by the RNAi machinery. To directly test the ability of 1360 to silence a nearby gene in vivo, we introduced a P element construct carrying a single copy of 1360 upstream of the hsp70-white reporter into flies. This 1360 element contributes to HP1-dependent variegation at a pericentric insertion site, as demonstrated by a decrease in silencing after FLP-mediated removal of 1360. In euchromatin, 1360 is not sufficient to induce silencing, suggesting that proximity to pericentric heterochromatin and/or a high local TE density contribute to heterochromatin formation. Silencing of the 1360, hsp70-white reporter is sensitive to mutations in RNAi components. Our results implicate 1360 as a target for sequence-specific heterochromatic silencing through an RNAi-dependent mechanism.
Results and Discussion
A Population of Sense and Antisense siRNAs Corresponding to 1360
In plants and yeast, RNAi and heterochromatin formation appear to be interconnected; the stability of heterochromatic marks at TEs depends upon components of the RNAi machinery [4-6]. While a different mechanism might apply to regions of satellite DNA, a parallel system might be found in domains with a high density of TEs in metazoa. To determine whether copies of the D. melanogaster TE 1360 (a.k.a. hoppel) might be targets of RNAi, we looked for evidence of 1360 transcription and processing. We carried out a pair-wise BLAST analysis (bl2seq, [7]) against the 4,480 bp ProtoP consensus sequence, the progenitor of 1360 derivatives [8], to visualize the composition of the subset of annotated 1360 copies that appear within a 200 kb region (chromosome four coordinates 300000-500000, Flybase Release 4.2) where we have mapped several silenced P reporter inserts (described in [3]). ProtoP contains a promoter [9] and an open reading frame (ORF) that is predicted to encode a transposase [8] (Figure 1A). The copies of 1360 analyzed here contain highly similar sequences within ∼1 kb of the terminal inverted repeat (TIR) ends, but the predicted ProtoP promoter and most of the ORF are deleted (Figure 1A). Additional transcription start sites are predicted by a study of the Drosophila Su(Ste) silencing system. A 1360 fragment within the Su(Ste) repeat of the Y chromosome has been implicated in the production of double-stranded RNA (dsRNA) and post-transcriptional silencing of Stellate. The transcription start sites that have been identified within the 1360 sequence of Su(Ste) are found in most of the 1360 elements from the chromosome four subset (Figure 1A) and in all of the 1360s that map within 10 kb of a silenced P reporter insert. These copies of 1360 may be competent to produce transcripts targeting dsRNA-mediated silencing to the region of chromatin in which they are embedded.
Figure 1.
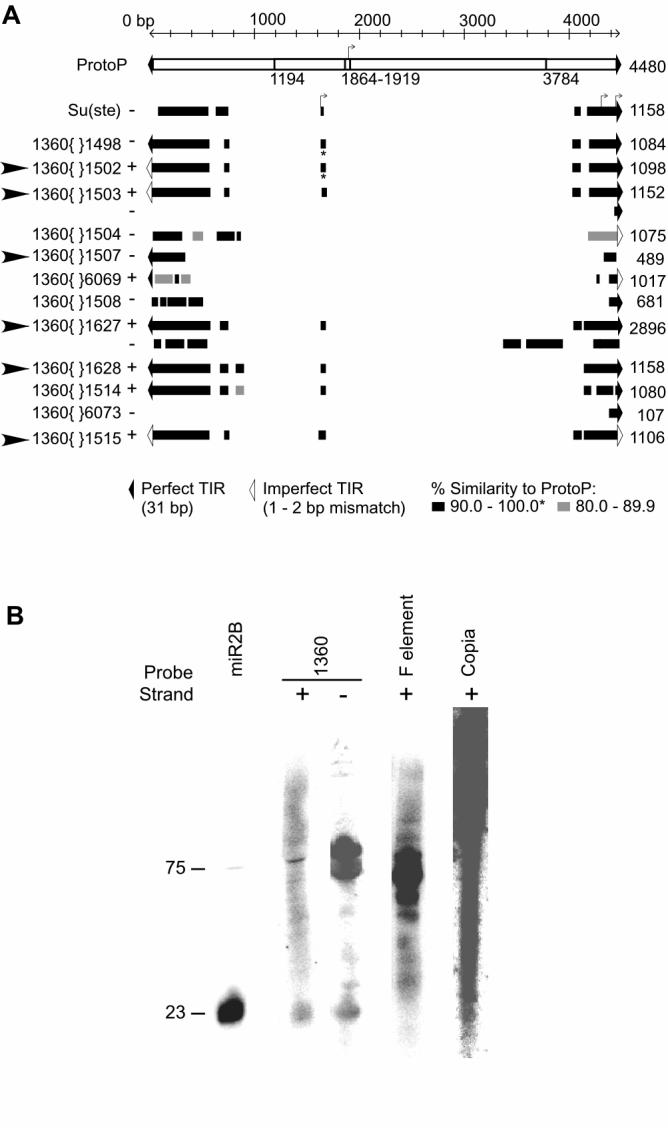
Short RNAs Correspond to Sense and Antisense Strands of 1360
(A) A 1360 fragment from the Su(Ste) repeat (pSY15.1) [18] and copies of 1360 annotated within coordinates 300000..500000 in region 102B of chromosome four (Flybase Release 4.2) compared to the ProtoP consensus [8]. ProtoP (white horizontal bar) contains an ORF that encodes transposase (1194-3784) [8] and a sequence identical (except for a single mismatch at 1897) to the predicted 1360 promoter (1869-1919) [9]; the predicted transcription start site is indicated by a bent arrow (1909) [9]. The Flybase identifier of each 1360 fragment is followed by + or -indicating the orientation of the homologous sequence (black and grey bars, total length shown at right) relative to ProtoP. Arrow heads (far left) indicate cases where a line has been recovered with a P reporter within 10 kb of the 1360; in all such cases, the reporter is variegating (described in [3]). The Su(Ste) sequence contains three putative transcription start sites within < 450 bp of the TIR [18]. The 1360 fragments have large internal deletions, but maintain highly conserved sequences near the ends; many show conservation of Su(Ste) transcription start sites.
(B) The siRNA profile of TEs from Drosophila Kc cells as shown by Northern blot. RNA of ca. 23 nt is observed using either the plus strand or minus strand of 1360{}1503 as a probe.
Small interfering RNAs (siRNAs) corresponding to heterochromatic TEs have been identified in yeast and plants [10, 11]. A previous cloning experiment has identified short RNAs that correspond to 38 different TEs (40% of the known elements), as well as satellite DNA and the subterminal minisatellite at the 2L telomere in D. melanogaster [12]. These small RNAs (16-28 nts) are most frequently found in testis and early embryos; the latter observation suggests a role in the early establishment of heterochromatin structure. Four of the small RNA clones recovered (17-25 nt) all correspond to the plus strand of 1360 [12]. To look for both plus and minus strand 1360 RNAs, we carried out strand-specific Northern blot analysis of small RNAs from Drosophila Kc cells (an embryonic cell line) using the genomic fragment 1360{}1503 as a probe. We detect a distinct population of small RNAs corresponding to both the sense and antisense strands of 1360 (Figure 1B). The size of these 1360 small RNAs is consistent with that of miR2B, a well characterized 23 nt siRNA that has been co-purified with RISC, suggesting processing by Dicer [13]. While a 23 nt band is not the dominant product for other TEs including the F element and copia, we observe small RNA species of different sizes. Expression of these TEs, including 1360, increases several fold in response to a knock-down of Dicer1/Dicer2 in Kc cells (Figure S1), arguing that they are directly or indirectly silenced by the RNAi machinery.
1360 Contributes to Silencing of an Adjacent Reporter
We used a P[hsp26-plant, hsp70-white] reporter to map over 20 sites within the distal arm of chromosome four that either allow full expression (indicative of euchromatic packaging) or induce position effect variegation (PEV, the mosaic silencing of a normally active gene placed in or near heterochromatin) of that reporter [3]. Analyzing a 200 kb region around the Hcf gene in detail, we found that reporters inserted less than 10 kb from sequences homologous to transposable element 1360 are generally variegating; this silencing depends upon Heterochromatin Protein 1 (HP1) [3]. In the two cases specifically tested [lines 39C-12 and 118E-10], silencing is also dependent on RNAi components including piwi, aubergine and homeless [14].
To directly test whether a single fragment of 1360 could function as a cis-acting silencer in vivo, we generated P[FRT-1360-FRT, hsp70-white], a mobile P construct carrying an FRT-flanked copy of 1360 upstream of the hsp70-white reporter gene (Figure 2, abbreviated “P[T1]”). Once this construct was introduced into flies, the silencing function of 1360 was assessed by the degree and frequency of hsp70-white transgene silencing. A total of 22 independent fly lines carrying a single P[T1] insert were recovered (1 variegated, 2 orange, 19 full red eye) by embryo microinjection and germ-line uptake followed by P element mobilization (described in Supplemental Experimental Procedures). We also created a fly line carrying P[hsp70-white] (abbreviated as “P[W]”) as a reference for maximum hsp70-white expression. The variegating phenotype of line T190-177 (located near the base of chromosome 2L) suggests that 1360 plays a role in heterochromatin formation in this case. To determine whether the adjacent 1360 is necessary for variegation of hsp70-white in this line, we carried out FLP-mediated excision of the 1360 in vivo (as described in Supplemental Experimental Procedures). Successful excision of 1360 at T190-177 (“-1360”, Figure 3A and 3B) leads to a significant suppression of PEV (loss of silencing) compared to sibling lines where 1360 failed to excise (“+1360”). More red pigment is present when two copies of the transgene are present; thus the residual silencing is best scored in hemizygous P/+ flies carrying a single unpaired copy of the P element. Quantitative eye pigment measurements from +1360 and -1360 hemizygotes (P/+) confirm that eye pigment is significantly greater in the -1360 hemizygotes. In agreement with these data, heat-shock induced hsp70-white transcript levels are more than three times higher in a line lacking 1360 (FLP1) than in the original line that carries an intact copy of 1360 (T190-177) (Figure 3C). Thus, a local copy of 1360 enhances silencing of percientric insert T190-177.
Figure 2.
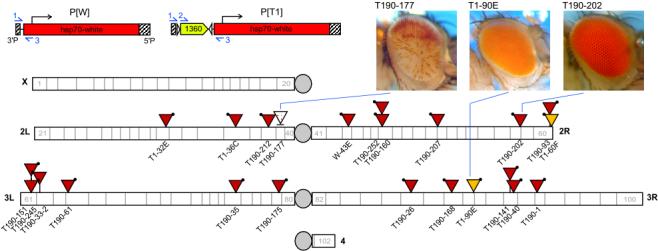
PInsertion Lines Generated by Microinjection and T1-90E Mobilization The silencing capacity of 1360 is assessed by white variegation in the transformant lines. Construct maps (upper left) show P[W], a reference for maximum hsp70-white expression, and P[T1] which contains a FRT-flanked 1360{}1503 placed upstream of hsp70-white. Slashed box = P element ends 3’P (left) and 5’P (right), white arrowheads = FLP recognition sequences (FRT), blue arrows = primers. Primers 1, 2 and 3 (see Supplemental Experimental Procedures for primer sequences) were used for PCR amplification of adult genomic DNA to determine the state of the transgenic 1360. The annotated euchromatic D. melanogaster genome (FlyBase Genome Browser Release 4.1, 2005) is shown (white rectangles = chromosome arms, numbers = polytene divisions, grey circles = centromeres). The P insertion site in each fly line (described in detail in Table S1) is marked by a triangle (red = full red eye, orange = orange eye, speckled = variegated eye, black dot = 5’P end). A horizontal line under T190-177 indicates its two putative map positions. Representative photos of variegating, orange and full red eye phenotypes are shown (top right). In most instances, the single copy of 1360 present in the reporter construct is not sufficient to induce silencing, but in one case (T190-177), a variegating phenotype is observed.
Figure 3.
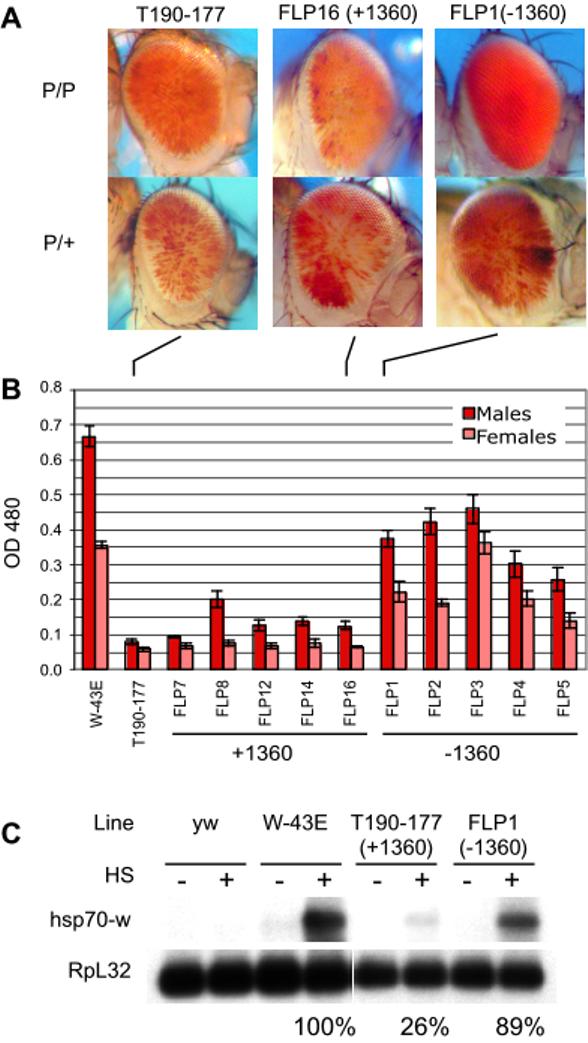
FLP-mediated Removal of 1360 Causes Loss of Silencing at T190-177
(A) Comparison of eye pigment levels in hemizygotes from +1360 vs. -1360 lines. FLP-mediated excision of 1360 is described in Supplemental Experimental Procedures. Total pigment levels are higher when two copies of the transgene are present.
(B) Quantitative measurement of eye pigmentation was carried out as described [3]. Bars represent the mean of triplicate samples (10 flies each) with standard error indicated (thin black line). white+ males typically show increased levels of red eye pigment compared to females. Lines lacking the 1360 element show increased expression.
(C) Northern blot analysis of hsp70-white transcripts. Numbers represent the means of triplicate white:RpL32 (signal:loading control) ratios normalized to W-43E (control line carrying P[W], set at 100 %). FLP-mediated excision of the 1360 element restores inducible expression from the silenced locus.
Most of the P[T1] inserts are fully active, as indicated by a full red eye phenotype (see Figure 2 and Table S1). Since loss or alteration of 1360 could have occurred during P element mobilization, we determined the state of the transgenic 1360 sequence using PCR (described in the legend to Figure 2). Our PCR analysis of P[T1] lines with red eyes did not detect any changes in 1360, indicating that the 1360 is present, but as a single copy is not sufficient to induce silencing of the reporter in these cases. Position of the P element within the genome might influence the ability of 1360 to induce silencing in cis. We generated a map of the P element insertion sites using inverse PCR to amplify sequences flanking the 5’P end of each insert for sequencing (described in the legend to Figure 2). Variegating insert T190-177 is flanked by a copy of invader4 within a TE-rich domain at the base of chromosome 2L at one of two possible insertion sites (42.8% or 81.5% TE sequence), while the fully active inserts generally map within TE-poor regions in euchromatin (Table S1). One exception is red-eyed insert T190-212, which maps to the right of a large block of TE fragments at region 38C. This region shows weak HP1 immunostaining on polytene chromosomes of various D. melanogaster strains [15] and relatively little HP1 or SU(VAR)3-9 binding in Kc cells [16], suggesting that this region does not represent an HP1-associated domain. In fact, T190-212 lies within a putative gene, which might be insulated from silencing.
The non-variegated reduction of hsp70-white expression in orange-eyed line T1-90E (carrying an insert within the stripe gene) suggests non-heterochromatic repression [17]. T1-60F, which carries a P element that maps near telomere associated sequence (TAS) repeats at the tip of chromosome 2R, exhibits a weak telomere position effect phenotype. Excision of 1360 from inserts T1-90E and T1-60F has no impact on eye phenotype (data not shown), suggesting that 1360 does not influence hsp70-white expression in these lines. In genomic environments outside the pericentric domain, the silencing function of 1360 appears to be inoperative or subordinate to other means of transcriptional regulation.
HP1-dependent Silencing of P[T1] is Sensitive to Mutations in the RNAi Machinery
PEV in line T190-177 is enhanced in the presence of increased HP1 dosage, and suppressed with decreased HP1 and SU(VAR)3-9 dosage, as is generally the case for reporters in pericentric heterochromatin (Figure 4). The presence of siRNAs corresponding to 1360 led us to investigate a role for the RNAi machinery in heterochromatin formation at 1360 by testing whether RNAi components are necessary for heterochromatic silencing of the P[T1] reporter in line T190-177. We observe that mutations in components of the RNAi pathway act as dominant suppressors of PEV in this case (Figure 4). Loss of the Argonaute family proteins PIWI and AUBERGINE (piwi1 and aubQC42, respectively) disrupts silencing at T190-177; these data point towards a requirement for RNA binding in transcriptional gene silencing. HOMELESS, a DEAD box helicase required for TE silencing [18], is also required. Deletions of the catalytic domains in the two Drosophila Dicer genes (dcr-1Q1447X and dcr-2R416X [19]) have a modest, but significant effect on PEV in this case. This genetic analysis reinforces previous observations that piwi, aub and hls are necessary for heterochromatin formation at reporters inserted near endogenous 1360s in D. melanogaster [14]. Here, we demonstrate that these RNAi components, plus Dicer and a cis-acting DNA element (1360) that generates siRNAs, contribute to HP1-dependent silencing. We investigated further by asking whether sensitivity of hsp70-white PEV in line T190-177 to these mutations is dependent upon the presence of the adjacent copy of 1360. Both +1360 and -1360 lines show a loss of silencing (Figure 4); thus both formation of the surrounding native heterochromatin and the local effect of 1360 appear to require HP1, SU(VAR)3-9 and components of the RNAi system. Normalization indicates that the impact of the additional 1360 is particularly dependent on HP1, SU(VAR)3-9 and AUBERGINE, suggesting that these components may promote the spread of nearby heterochromatin into the reporter gene.
Figure 4.
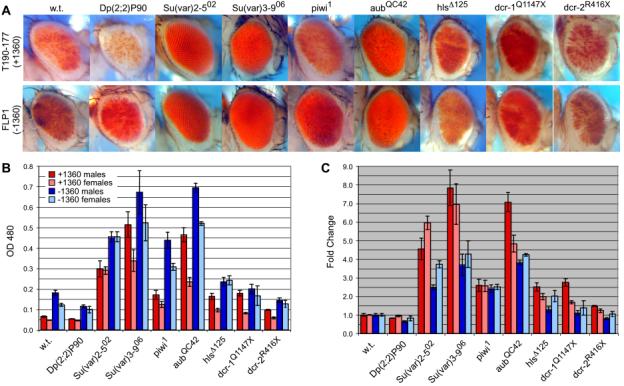
HP1-dependent Silencing at T190-177 is Sensitive to Mutations in the RNAi MachineryPhotos (A) and eye pigment levels (B) of male progeny from a cross between males carrying either variegating insert T190-177 (+1360) or FLP1 (indentical except for the excision of 1360) and the following females: yw67c23, wild type (“w.t.”); Su(var)2-502, HP1 loss of function; Dp(2;2)P90, HP1 duplication; Su(var)3-906, HMTase null; piwi1, aubQC42, hlsD125, mutations in RNAi components; dcr-1Q1147X, Dicer-1 loss of function; dcr-2R416X, Dicer-2 loss of function. (C) Each bar represents the mean of triplicate values normalized to the mean wild type eye pigment value for T190-177 (red and pink) or FLP1 (blue and light blue). Silencing at this locus is dependent upon HP1, SU(VAR)3-9 and the RNAi machinery.
Conclusions
Fine scale mapping within a 200 kb interval in region 102B of D. melanogaster chromosome four led us to identify fragments of 1360 as putative signals for heterochromatin recruitment [3]. Here, we have directly tested a role for 1360 sequences in heterochromatin formation. We find that a single copy of 1360 contributes to HP1- and SU(VAR)3-9-dependent silencing of an adjacent gene within a TE-rich environment. In contrast, a single copy of 1360 has no impact on reporter gene expression at loci outside of the pericentric domain. This suggests that the local density of repeats influences heterochromatin formation. A survey of HP1 binding in Drosophila cells shows that TEs tend to be bound by HP1 when flanked by additional repeats [20]. The single copy of 1360 in our test construct apparently facilitates spreading of heterochromatin complexes to encompass the hsp70-white transgene, but by itself is insufficient to generate stable heterochromatic packaging. Spatial positioning within the genome also influences heterochromatic silencing at TEs. Translocation of the entire banded arm of D. melanogaster chromosome four away from the centromere diminishes silencing of a fourth chromosome reporter [21]. The variegating P element insert in line T190-177, which lies within a repeat-rich proximal domain close to the centromere, does not allow us to discriminate between these two parameters. However, the presence of euchromatic domains on the small fourth chromosome [3, 22] argues that proximity to the centromere per se is not sufficient to induce silencing, pointing once again to the role of local determinants.
Currently, no data supports an inherent capacity for heterochromatin proteins to specifically bind TE sequences, making a model of RNAi-directed silencing attractive. We observe ∼23 nt RNAs complementary to sense and antisense strands of 1360. The presence of sequences from both strands suggests that these small RNAs originate from a dsRNA precursor; their size is consistent with known Dicer products such as miR2B (Figure 1B). Furthermore loss of Dicer, an enzyme required for dsRNA processing, leads to loss of PEV (Figure 4) and loss of 1360 silencing (Figure S1). The 1360 short RNAs we observe by Northern blot could function in targeting transcriptional silencing to 1360 DNA sequences within heterochromatic regions via a RISC-like ribonucleoprotein complex. In S. pombe, the RISC-like RNA-induced Initiation of Transcriptional Silencing (RITS) complex contains an Argonaute family member (Ago1) and siRNAs homologous to the heterochromatic dg-dh repeats [23]. The genetic analyses presented here demonstrate that silencing at P[T1] depends upon the Argonaute family proteins AUBERGINE and PIWI. HOMELESS, a DEAD box helicase that is required for the repression of TE transcription [18] also has an impact on this HP1-dependent silencing. At insert T190-177, silencing is disrupted by mutations in RNAi components both in the presence and absence of 1360, indicating that silencing mediated by both the local element and the surrounding heterochromatin depends upon the RNAi system. While this leaves open the question of whether RNAi acts directly upon 1360, the presence of 1360 small RNAs suggests that this element is a direct target of the RNAi machinery.
Whether the siRNAs are generated locally, or derive from transcription elsewhere (as can occur in plants and in C. elegans [6, 24]) is not known for this system. In D. melanogaster, transcripts from within the ORF of 1360 (hoppel) can be detected via RT-PCR [9]. The 1360 fragments we have analyzed contain conserved sequences (Figure 1A) that could serve as recognition sites as well as sequences corresponding to the Su(Ste) 1360 transcription start sites previously detected by 5’-RACE and primer extension mapping [18]. Whether the copies of 1360 on chromosome four or at T190-177 are actually transcribed has yet to be determined. The results reported thus far provide initial insights into a role for TEs in sequence-specific heterochromatic silencing in D. melanogaster and support a role for RNAi in this process.
Supplementary Material
Acknowledgements
Special thanks to R. Carthew (Northwestern University) for Dicer mutant stocks, J.C. Eissenberg (Saint Louis University) for stock Dp(2;2)P90, and to W. Leung for assistance with the pair-wise BLAST analysis. This work was supported by grant GM068388 to S.C.R. Elgin from the National Institutes of Health (NIH). K.A. Haynes was supported in part by the Washington University Chancellor’s Fellowship. A.A. Caudy was supported in part by a predoctoral fellowship from the Howard Hughes Medical Institute (HHMI).
References
- 1.Haynes KA, Leibovitch BA, Rangwala SH, Craig C, Elgin SC. Analyzing heterochromatin formation using chromosome 4 of Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 2004;69:267–272. doi: 10.1101/sqb.2004.69.267. [DOI] [PubMed] [Google Scholar]
- 2.Elgin SC, Grewal SI. Heterochromatin: silence is golden. Curr Biol. 2003;13:R895–898. doi: 10.1016/j.cub.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Sun FL, Haynes K, Simpson CL, Lee SD, Collins L, Wuller J, Eissenberg JC, Elgin SC. cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol Cell Biol. 2004;24:8210–8220. doi: 10.1128/MCB.24.18.8210-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 5.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 6.Matzke M, Aufsatz W, Kanno T, Daxinger L, Papp I, Mette MF, Matzke AJ. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim Biophys Acta. 2004;1677:129–141. doi: 10.1016/j.bbaexp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 7.Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 8.Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci U S A. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiss D, Quesneville H, Nouaud D, Andrieu O, Anxolabehere D. Hoppel, a P-like element without introns: a P-element ancestral structure or a retrotranscription derivative? Mol Biol Evol. 2003;20:869–879. doi: 10.1093/molbev/msg090. [DOI] [PubMed] [Google Scholar]
- 10.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 11.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 12.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 13.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 15.Fanti L, Berloco M, Piacentini L, Pimpinelli S. Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: a cytological map of euchromatic HP1 binding sites. Genetica. 2003;117:135–147. doi: 10.1023/a:1022971407290. [DOI] [PubMed] [Google Scholar]
- 16.Greil F, van der Kraan I, Delrow J, Smothers JF, de Wit E, Bussemaker HJ, van Driel R, Henikoff S, van Steensel B. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev. 2003;17:2825–2838. doi: 10.1101/gad.281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 18.Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 19.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 20.de Wit E, Greil F, van Steensel B. Genome-wide HP1 binding in Drosophila: developmental plasticity and genomic targeting signals. Genome Res. 2005;15:1265–1273. doi: 10.1101/gr.3198905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cryderman DE, Morris EJ, Biessmann H, Elgin SC, Wallrath LL. Silencing at Drosophila telomeres: nuclear organization and chromatin structure play critical roles. Embo J. 1999;18:3724–3735. doi: 10.1093/emboj/18.13.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun FL, Cuaycong MH, Craig CA, Wallrath LL, Locke J, Elgin SC. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc Natl Acad Sci U S A. 2000;97:5340–5345. doi: 10.1073/pnas.090530797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


