Abstract
Heat shock protein 70 (Hsp70) is a well-known inhibitor of apoptotic pathways; however, a role for Hsp70 in the modulation of death receptor–mediated apoptosis remains largely unexplored. In this study, the ability of Hsp70 to modulate tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-induced apoptosis was examined in SW480 and CCRF-CEM cells. These lines exhibit the characteristics of type I cells (SW480, human colon adenocarcinoma), with no requirement for mitochondrial involvement to exhibit apoptosis following death receptor engagement and type II cells (CCRF-CEM, human leukemic T cell), which do require amplification of the signal through the mitochondria. Unexpectedly, expression of Hsp70 in the type II CCRF-CEM cells enhanced the extent of TRAIL-induced apoptosis, but in SW480, Hsp70 had no impact on TRAIL-induced apoptosis. The enhanced TRAIL-induced apoptosis was accompanied by an up-regulation of TRAIL receptors, R1 and R2, at the cell surface as determined by flow cytometry and at the transcriptional level as assessed by real-time polymerase chain reaction (PCR). Increased expression of Hsp70 led to up-regulated expression of p53, and chromatin immunoprecipitation combined with real-time PCR revealed increased binding of p53 to its consensus sequence in the TRAIL-R2 gene. In contrast, expression of Hsp70 in SW480 cells did not increase p53 or TRAIL-R1 or TRAIL-R2 surface expression. This result is in marked contrast to most apoptotic stresses, including TNFα and Fas ligand, where Hsp70 has been shown to inhibit apoptosis in type II cells. These findings suggest that in tumors retaining functional p53 and expressing high levels of Hsp70, TRAIL may be an effective therapy.
INTRODUCTION
Apoptosis is a tightly regulated and genetically controlled event crucial to normal development and tissue homeostasis (Krammer 1999; Vaux and Korsmeyer 1999). Aberrations in the control of apoptosis can lead to a number of physiological disorders including cancer, in which apoptosis is often disrupted, thus conferring a survival advantage to the tumorigenic cells (Hanahan and Weinberg 2000; Green and Evan 2002). Apoptosis can be divided into 2 distinct but interconnecting pathways: the extrinsic pathway activated upon ligation of death receptors of the tumor necrosis factor (TNF) receptor superfamily and the intrinsic pathway, which is initiated by cellular stresses that activate proapoptotic members of the Bcl-2 family to target the mitochondria. Central to both pathways are the caspases, which cleave a specific set of target substrates leading to the classic hallmarks of apoptosis (Thornberry and Lazebnik 1998). Activation of the apical caspases, caspase-8 and caspase-10 in the extrinsic pathway, is mediated by the adaptor protein Fas-associated death domain (FADD) through formation of the death-inducing signaling complex (DISC) at the cytoplasmic death domains of ligated death receptor oligomers (Kischkel et al 1995; Medema et al 1997; Kischkel et al 2001). In a similar fashion, the initiator caspase in the intrinsic pathway, caspase-9, is activated at the apoptosome complex, which forms upon stress-induced release of cytochrome c from the mitochondria (Li et al 1997; Zou et al 1997). In both pathways, activation of apical caspases initiates a cascade of caspase activation leading to apoptosis. Cross talk between the extrinsic and intrinsic pathways exists through caspase-8–mediated cleavage of the proapoptotic Bcl-2 protein, Bid (Li et al 1998; Luo et al 1998). Truncated Bid activates the proapoptotic molecules Bax and Bak, which target the mitochondria and initiate the intrinsic pathway (Eskes et al 2000; Wei et al 2000, 2001). In some cells, called type II cells, recruitment of the intrinsic pathway is required for efficient apoptosis and can be inhibited by Bcl-2 (Scaffidi et al 1998), although this concept is controversial (Huang et al 1999, 2000; Schmitz et al 1999). In type I cells, caspase-8–mediated activation of downstream effector caspases is sufficient to trigger apoptosis without mitochondrial involvement.
TNF-related apoptosis-inducing ligand (TRAIL) induces apoptosis through ligation of 1 of the 2 cognate receptors that contain intracellular death domains, TRAIL-R1 or TRAIL-R2 (Almasan and Ashkenazi 2003). Interest in TRAIL as a cancer therapy developed after demonstration that TRAIL can selectively induce apoptosis in tumor cells both in vivo and in vitro, whereas normal cells remain refractory (Wiley et al 1995; Pitti et al 1996; Ashkenazi et al 1999; Walczak et al 1999; Kelley et al 2001). In addition, TRAIL in combination with certain deoxyribonucleic acid (DNA)-damaging drugs or radiotherapy shows synergistic antitumor effects (Ashkenazi et al 1999; Bonavida et al 1999; Gliniak and Le 1999; Chinnaiyan et al 2000; Nagane et al 2000). In some cases, this may be due to p53-mediated up-regulation of TRAIL-R1 or TRAIL-R2 (Sheikh et al 1998; Wu et al 2000; Guan et al 2001; Arizono et al 2003). However, many cancer cell lines remain resistant to TRAIL-induced apoptosis, possibly due to the expression of 2 decoy receptors for TRAIL, TRAIL-R3 and TRAIL-R4 (Ashkenazi 2002).
The heat shock proteins (Hsps) are a family of highly conserved and abundantly expressed proteins. While acting as molecular chaperones in unstressed cells, Hsps promote cell survival during periods of both acute and chronic stress (Lindquist 1986). The ability of cells to develop thermotolerance, a state of transient resistance to severe stress, after a mild heat shock is thought to be largely the responsibility of Hsp70 (Subjeck et al 1982). In addition, expression of Hsp70 alone has been shown to inhibit apoptosis induced by a variety of stresses including TNFα, heat, ultraviolet radiation, ceramide, and a number of cytotoxic drugs (Jaattela et al 1992; Samali and Cotter 1996; Gabai et al 1997; Mosser et al 1997; Buzzard et al 1998; Steel et al 2004; Guo et al 2005). The significance of these findings is highlighted by studies that correlate high expression of Hsp70 in tumors with resistance to standard cancer therapies, increased metastasis to lymph nodes, and a poor prognosis for disease-free status and overall survival (Ciocca et al 1993; Jaattela 1999; Creagh et al 2000; Jolly and Morimoto 2000). The oncogenic activity of Hsp70 stems from its ability to protect against protein damage, stabilize mutant protein conformations, and inhibit apoptotic pathways (Mosser and Morimoto 2004).
We and others have demonstrated that Hsp70 can inhibit TNFα-induced apoptosis (Jaattela et al 1992; Buzzard et al 1998). In addition, we recently reported that Hsp70 inhibits Fas-mediated apoptosis upstream of the mitochondria in type II cells (Clemons et al 2005). In this study, we investigated the ability of Hsp70 to modulate TRAIL-induced apoptosis in a type I cell line, SW480, and a type II cell line, CCRF-CEM. We found that neither Hsp70 nor Bcl-2 could inhibit TRAIL-induced apoptosis in SW480 cells, confirming that Hsp70 cannot modulate the extrinsic apoptosis pathway. Surprisingly, although CCRF-CEM cells were refractory to TRAIL, expression of Hsp70 sensitized them to TRAIL by a mechanism involving p53-mediated up-regulation of TRAIL-R1 and TRAIL-R2. These results add to the pleiotropic effects that Hsp70 has on apoptotic pathways and suggests that tumors expressing high levels of Hsp70 and that retain functional p53 may be good targets for therapy with TRAIL.
MATERIALS AND METHODS
Reagents and antibodies
Recombinant human TRAIL was from Genentech (South San Francisco, CA, USA) and is described elsewhere (Ashkenazi et al 1999; Lawrence et al 2001). The pancaspase inhibitor z-VAD-fmk was from Biomol (Plymouth Meeting, PA, USA). Annexin-V-FLUOS was from Roche Diagnostics (Mannheim, Germany). The following antibodies were used: human caspase-8 antibody (clone C15) (kind gift from Prof. P. Krammer, German Cancer Research Center, Heidelberg, Germany); human Bid antibody (Cell Signaling Technology, Beverly, MA, USA); human caspase-9 and caspase-7 antibodies (kind gifts from Dr. Y. Lazebnik, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA); p53 (clone DO-1), MDM2 (clone SMP14), Bax (clone 6A7), and human caspase-3 (clone 19) antibodies (BD Biosciences, San Diego, CA, USA); human Hsc70 antibody (clone IB5) (StressGen Biotechnologies, Victoria, British Columbia, Canada); human Hsp70 antibody (clone N15) (kind gift from Dr. W. Welch, University of California, San Francisco, CA, USA); β-tubulin antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA); fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (Zymed Laboratories, South San Francisco, CA, USA); human TRAIL-R1 (clone M271), TRAIL-R2 (clone M412), TRAIL-R3 (clone M430), and TRAIL-R4 (clone M444) antibodies (Griffith et al 1999) (Amgen, Seattle, WA, USA); and mouse IgG1 and IgG2a immunoglobulins (DakoCytomation, Glostrup, Denmark).
Cell lines and stable transfections
The human leukemic T cell line CCRF-CEM and the colon adenocarcinoma cell line SW480 were maintained in RPMI 1640 (Invitrogen Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal calf serum and 1% penicillin-streptomycin. All cultures were maintained at 37°C in a humidified 5% CO2 atmosphere and were routinely monitored for mycoplasma contamination.
CCRF-CEM cells were transfected by electroporation with the mammalian expression vector pCI-neo containing the coding region of the major human inducible hsp70 gene, HspA1A, derived from pH2.3 (a kind gift from Dr. C. Hunt, Washington University, St Louis, MO, USA). Stable cells expressing hsp70 were selected using 700 μg/ mL G418 (Invitrogen Life Technologies). Transfected cells were single-cell cloned, and expression of Hsp70 in clones, parental cells, and vector control cells was analyzed by Western blotting.
SW480 cells were transfected with the retroviral plasmid MSCV-IRES-GFP containing human hsp70 (derived from pH2.3) or the mammalian expression vector pMPZenSVNeo containing a human Bcl-2 construct (a kind gift from Dr. A. Strasser, Walter and Eliza Hall Institute, Melbourne, Australia) using Lipofectamine 2000 (Invitrogen Life Technologies). A population of cells expressing hsp70 was isolated by fluorescence-activated cell sorter (FACS) sorting (BD Biosciences FACStar) for green fluorescent protein (GFP) expression. GFP expression of vector control cells was matched with that of hsp70 transfected cells. Cells stably expressing Bcl-2 were selected using 1 mg/mL G418 (Invitrogen Life Technologies). Expression of Hsp70 and Bcl-2 was confirmed by Western blotting.
Cell treatments
All experiments using SW480 cells were performed on approximately 50–70% confluent cell cultures, whereas all experiments using CCRF-CEM cells were performed on cultures at a density of 1 × 106 cells/mL. Immediately prior to each experiment, the medium was replaced with fresh, pH-equilibrated medium. TRAIL was diluted in serum-free medium to appropriate concentrations immediately before use and added to cells for 18 hours unless indicated otherwise. Genentech TRAIL does not require cross-linking to induce apoptosis (Ashkenazi et al 1999; Lawrence et al 2001). For some experiments, cells were pretreated for 1 hour with 50 μM z-VAD-fmk dissolved in dimethyl sulfoxide (DMSO). An equivalent volume of DMSO (final concentration, 0.2%) was added to control samples. Cells were irradiated using a cesium source. Heat shock was achieved by full immersion of flasks of cells in a circulating water-bath calibrated to 44 ± 0.1°C for 15 to 60 minutes.
Apoptosis and long-term survival assays
Apoptosis was assessed either by labeling with Annexin-V-FLUOS or by scoring nuclear morphology. Annexin-V-FLUOS labeling was performed according to the manufacturer's instructions (Roche Diagnostics), and cells were analyzed by flow cytometry on a BD Biosciences FACSCalibur. Ten thousand cells were counted for each sample.
Nuclear morphology was assessed by fluorescence microscopy following propidium iodide staining as described previously (Buzzard et al 1998). A cell was scored as apoptotic if it displayed one or more of the following: nuclear margination, chromatin condensation, or formation of apoptotic bodies. At least 200 cells were counted in each experiment.
Long-term survival of CCRF-CEM cells was assessed by clonogenic assay. A series of dilutions containing 200 μL of known numbers of cells (5000 to 0.5 cells/mL) were prepared, placed into 96-well U-bottom plates, and incubated at 37°C for 10–12 days. Wells were scored as positive or negative for colony growth as described previously (Furth et al 1981).
Immunoblotting
Samples containing equal amounts of protein were separated by 13% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Primary and secondary antibodies were added in 1% milk powder in phosphate-buffered saline (PBS) and incubated for 2 to 6 hours at room temperature or overnight at 4°C. Horseradish peroxidase– conjugated and alkaline phosphatase–conjugated secondary antibodies were detected by enhanced chemiluminescence (Lumilight; Roche Diagnostics), or colorimetric staining with naphthol phosphate (Sigma-Aldrich, St Louis, MO, USA) and fast red TK (Bio-Rad Laboratories, Hercules, CA, USA), respectively.
Immunofluorescence detection of TRAIL receptors
Cells were immunostained with 0.5 μg of TRAIL-R1 (IgG2a), TRAIL-R2 (IgG1), TRAIL-R3 (IgG1), or TRAIL-R4 (IgG1) antibody or isotype control immunoglobulins per 1 × 106 cells followed by 0.5 μg of FITC-conjugated anti-mouse antibody. Cells were analyzed by flow cytometry on a FACSCalibur. Ten thousand cells were counted for each sample.
SYBR Green real-time quantitative polymerase chain reaction
Total ribonucleic acid (RNA) was harvested from cells in log-phase of growth using TRIzol (Invitrogen Life Technologies) according to manufacturer's instructions. A standard 2 μg of RNA was reverse transcribed with a poly(dT) anchored primer using Superscript II reverse transcriptase (Promega, Madison, WI, USA).
Real-time quantitative polymerase chain reaction (RTQPCR) was performed on an ABI Prism 7000 Sequence Detection System using SYBR Green I chemistry (Applied Biosystems, Foster City, CA, USA). PCR consisted of approximately 20 ng of complementary DNA, 2 μM forward and reverse primers, and 2× SYBR Green I master mix reagent in a total reaction volume of 20 μL. The following primers were used: TRAIL-R1, forward, 5′-GATCGATGTGGTCAGAGCTGG-3′, and reverse, 5′-TGTGGATCGAGGCGTTCC-3′; TRAIL-R2, forward, 5′-GTGCCCTTGACTCCTGGG-3′, and reverse, 5′-AAGGTGTCCCTGTGGCCC-3′; TRAIL-R3, forward, 5′-CAGTTCCTCCCATCTTCAGGC-3′, and reverse, 5′-GGCTGAATAAATCCCGTGACC-3′; TRAIL-R4, forward, 5′-CTCCTACAAAGGGAAGCAGCC-3′, and reverse, 5′-CTAGGACCATTGGTAAGCTGCC-3′; p53, forward, 5′-CTTACTGCAGCCTTTGCCTCC-3′, and reverse, 5′-TGCAAAAGTTGGCTGGCC-3′; vimentin, forward, 5′-AGAGAACTTTGCCGTTGAAGCT-3′, and reverse, 5′-GAAGGTGACGAGCCATTTCC-3′. There are 2 splice variants for TRAIL-R2, and the primers were designed to detect both transcripts.
The following cycling method was employed: 50°C for 2 minutes, 95°C for 10 minutes, and then 40 cycles of 95°C for 15 seconds, 60°C for 60 seconds. Amplification of a target gene was compared with that of the housekeeping gene vimentin. Results are presented as relative transcript abundance (RTA) according to the following formula: RTA = 105/2(CtGENE − CtVIMENTIN), where Ct is the cycle number at which the PCR signal reaches the designated threshold value.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using an assay kit (Upstate, Charlottesville, VA, USA). For each sample, 1 × 107 cells were fixed in fresh PBS/1% formaldehyde solution and sonicated on ice with a Branson Sonifier at 30% power output and 3 duty settings for 4 rounds of 15 seconds. This sonication program generated a DNA smear with an average size of approximately 300 bp and ranging from 650 to 100 bp. All samples were precleared with 40 μL of protein A–agarose saturated with salmon sperm DNA by rotation at 4°C for 1 hour followed by immunoprecipitation with 5 μg of p53 antibody by rotation at 4°C overnight. Immune complexes were collected with 60 μL of protein A–agarose for 1 hour at 4°C with rotation, and immunoprecipitated DNA was analyzed by SYBR Green RTQPCR as described above using the following primers: p53 binding site, forward, 5′-CGCTCCACTTGGTGTAAATTCC-3′, and reverse, 5′-GAATTCCCTCCTTGTCGCC-3′; nonspecific control site, forward, 5′-GGTTCGATGGTCTTTTGGCC-3′, and reverse, 5′-TACTCCCACCTCAGCCTCCC-3′. Results are presented as the percentage of specific amplification from immunoprecipitated samples (CtIMM) compared to total input DNA (CtINPUT) according to the following equation: (1010/2CtIMM)/(1010/2CtINPUT) × 100%.
RESULTS
Effect of Hsp70 on TRAIL-induced apoptosis of SW480 and CCRF-CEM cells
The effect of Hsp70 on TRAIL-induced apoptosis, which uses a similar signaling pathway to that mediated through Fas, was examined. We demonstrated recently that Hsp70 inhibits Fas-mediated apoptosis of the type II cell line, CCRF-CEM, but not the type I cell line, SW480 (Clemons et al 2005). CCRF-CEM and SW480 cells were transfected with constructs containing the coding region of human hsp70 (Clemons et al 2005), and expression of Hsp70 in transfected cells was confirmed by Western blotting, as shown previously (Clemons et al 2005). Hsp70 transfected CCRF-CEM clones 7 and 16 expressed high levels of Hsp70, whereas clone 18 had similar levels of Hsp70 to parental or vector-only transfected cells (Clemons et al 2005). A bulk population of SW480 cells was generated that expressed high levels of Hsp70 compared to vector control or parental cells. The functionality of Hsp70 in transfected CCRF-CEM clones and SW480 cells was demonstrated previously by its ability to provide long-term survival after heat, as described elsewhere (Clemons et al 2005).
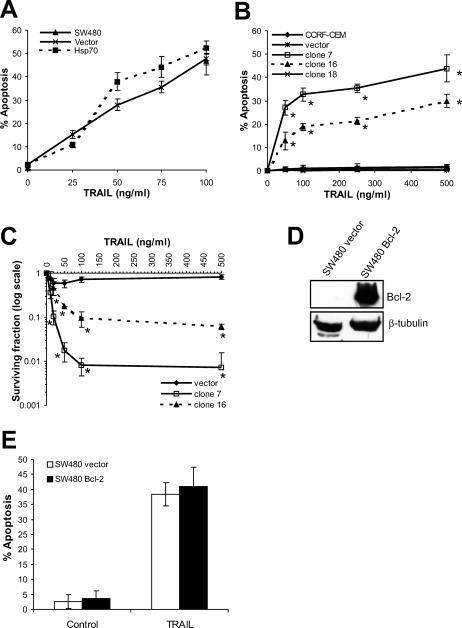
Cells were incubated with recombinant human TRAIL for 18–20 hours (Fig 1A,B). TRAIL induced apoptosis in SW480 cells in an Hsp70-independent manner. Small increases in apoptosis in Hsp70-expressing cells compared to vector-only transfected cells at 50 and 75 ng/mL TRAIL were not statistically significant (P > 0.05) (Fig 1A). TRAIL-induced apoptosis of parental SW480 cells was examined only at 100 ng/mL and is represented by a single point (Fig 1A). Although parental CCRF-CEM cells underwent Fas-induced apoptosis (Clemons et al 2005), they were refractory to the effects of TRAIL. Unexpectedly, expression of Hsp70 induced sensitivity to TRAIL (Fig 1B). When apoptosis was measured by labeling with Annexin-V-FLUOS and flow cytometry, both Hsp70-expressing clones demonstrated sensitivity to TRAIL at doses of 50 ng/mL and above, whereas parental, vector control and clone 18 cells were resistant to TRAIL at doses of up to 500 ng/mL. Compared to clone 16, clone 7 cells were more sensitive to TRAIL, consistent with their slightly higher levels of Hsp70 (Clemons et al 2005).
Fig 1.
Effect of Hsp70 expression on TRAIL-induced apoptosis of SW480 and CCRF-CEM cells. The effect of Hsp70 expression on TRAIL-induced apoptosis was examined in SW480 (A) and CCRF-CEM cells (B). Cells were incubated with the indicated doses of recombinant human TRAIL for 18–20 hours at 37°C, and apoptosis was assessed by scoring nuclear morphology (SW480) or by labeling with Annexin-V-FLUOS and flow cytometry (CCRF-CEM). The effect of TRAIL on parental SW480 cells was examined at a single dose (100 ng/mL). (C) Long-term survival was assessed by a clonogenic assay following 16-hour incubation at 37°C with TRAIL. Colony growth was assessed 10–12 days after plating. (D) Expression of Bcl-2 in transfected SW480 cells was compared with vector control cells by Western blotting. The effect of Bcl-2 on TRAIL-induced apoptosis in SW480 cells was examined (E). All graphs show means and standard deviations of at least 3 independent experiments. Statistical analysis by Student's t-test; *P < 0.05 compared to vector control
Long-term survival after incubation with TRAIL, as assessed by clonogenic assay, demonstrated that Hsp70 is not simply accelerating TRAIL-induced apoptosis in CCRF-CEM cells (Fig 1C). Significant differences in long-term survival were detected after incubation with doses of TRAIL of 25 ng/mL or higher. After incubation with 100 ng/mL TRAIL, clones 7 and 16 showed 100- and 10-fold reductions, respectively, in colony formation compared to vector control cells, which remained refractory to TRAIL in this long-term assay.
It has been reported previously that thermotolerance does not inhibit TRAIL-induced apoptosis of type I SW480 cells (Ozoren and El-Deiry 2002). This was attributed to the inability of Hsp70 or Hsp90 to inhibit the extrinsic apoptosis pathway. We examined the effect of Bcl-2 expression on TRAIL-induced apoptosis of SW480 cells. Overexpression of Bcl-2 in SW480 compared to vector control cells was confirmed by Western blotting (Fig 1D) but did not result in inhibition of TRAIL-induced apoptosis of SW480 cells (Fig 1E), indicating that recruitment of the intrinsic pathway is not required in these cells. Thus, our results are in agreement with previous findings that thermotolerant type I cells with increased levels of Hsp70 are not protected from TRAIL-induced apoptosis (Ozoren et al 2002). Because neither Bcl-2 nor Hsp70 can inhibit TRAIL-induced apoptosis in SW480 cells, it suggests that, like Bcl-2, Hsp70 cannot modulate the extrinsic pathway.
TRAIL activates the caspase cascade in Hsp70-expressing CCRF-CEM cells
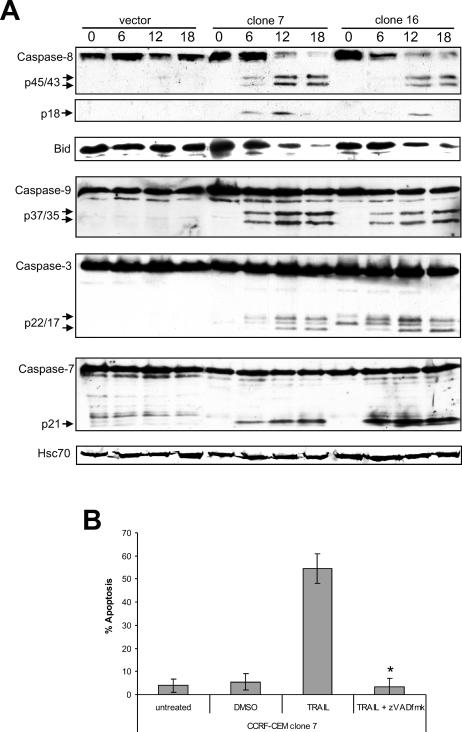
To elucidate the mechanism by which Hsp70 sensitizes CCRF-CEM cells to TRAIL, caspase cleavage was evaluated by Western blotting (Fig 2A). Cleavage of the initiator caspases, caspase-8 and caspase-9, as well as Bid and the downstream effector caspases, caspase-3 and caspase-7, was examined. No caspase cleavage was detected in vector control lysates in response to TRAIL, indicating that the mechanism of TRAIL resistance in these cells is at the level of, or upstream of DISC formation. In contrast, Hsp70-expressing cells exhibited extensive caspase cleavage after only 6-hour incubation with TRAIL. Cleavage of the initiator caspases, caspase-8 and caspase-9, and the effector caspases, caspase-3 and caspase-7, was detected. This indicated that Hsp70 was acting upstream of caspase-8 to sensitize these cells to TRAIL. Although the cleavage product of Bid could not be detected with this antibody, a decrease in the amount of full-length Bid was observed in Hsp70-expressing cells but not vector control cells (Fig 2A).
Fig 2.
TRAIL activates the caspase cascade in Hsp70-expressing CCRF-CEM cells. (A) Cells were incubated at 37°C with 100 ng/mL TRAIL for 0, 6, 12, or 18 hours prior to harvest. Equal amounts of protein were separated on a 13% SDS-PAGE gel and transferred to nitrocellulose membranes. Membranes were probed with specific antibodies that detect both the proform and cleavage products of human caspase-8, caspase-9, caspase-7, or caspase-3, or Bid. (B) Clone 7 cells were preincubated for 1 hour with 50 μM z-VAD-fmk in DMSO or an equivalent concentration of DMSO, and then incubated with 100 ng/mL TRAIL for 18 hours at 37°C. Apoptosis was evaluated by scoring nuclear morphology. The means and standard deviations of 3 independent experiments performed in duplicate are shown. Statistical analysis by Student's t-test; *P < 0.05 vs TRAIL alone
Preincubation with the pancaspase inhibitor z-VAD-fmk completely inhibited TRAIL-induced apoptosis in clone 7 cells (Fig 2B), confirming that TRAIL-induced apoptosis is caspase dependent in these cells.
TRAIL-R1 and TRAIL-R2 surface expression and messenger RNA are up-regulated in CCRF-CEM cells expressing Hsp70
Caspase-8 is the apical caspase in the extrinsic pathway and is activated upon formation of the DISC (Medema et al 1997). Thus, we postulated that Hsp70 could be inducing sensitivity to TRAIL in CCRF-CEM cells either by stabilizing the DISC, or by altering expression of TRAIL receptors either at a transcriptional level or through posttranslational mechanisms. There are 4 membrane-bound receptors for TRAIL; TRAIL-R1 and TRAIL-R2 are death-inducing receptors, whereas TRAIL-R3 and TRAIL-R4 are unable to induce apoptosis and are thought to be decoy receptors (Ashkenazi 2002).
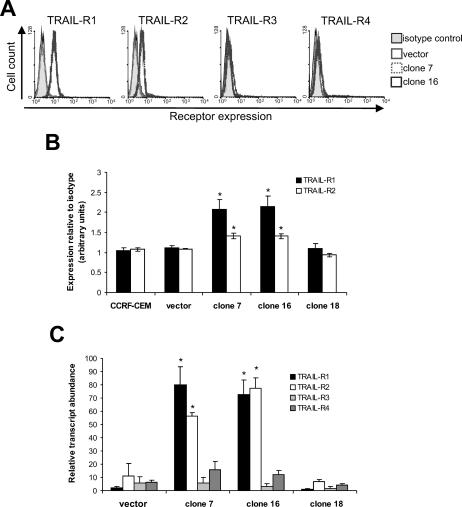
FACS analysis could not detect expression of any of the 4 receptors on parental and vector control cells, which accounts for their resistance to TRAIL (Fig 3A). In contrast, clones expressing Hsp70 had up-regulated both TRAIL-R1 and TRAIL-R2, but not TRAIL-R3 or TRAIL-R4 on the cell surface, thus providing an explanation for their sensitivity to TRAIL. To provide a quantitative comparison of the expression of TRAIL-R1 and TRAIL-R2, the expression of each receptor was compared with background binding of isotype control immunoglobulins from 3 separate experiments (Fig 3B). Both TRAIL-R1 and TRAIL-R2 were significantly up-regulated on Hsp70-expressing clones 7 and 16 compared to vector control and clone 18 cells (P < 0.05).
Fig 3.
TRAIL-R1 and TRAIL-R2 are up-regulated in CCRF-CEM cells expressing Hsp70. (A) Expression of TRAIL receptors on the plasma membrane was analyzed by flow cytometry. (B) For quantitation, results were expressed as the ratio of the geometric mean of receptor expression to the geometric mean of the corresponding isotype control. The means and standard deviations of 3 independent experiments are shown. (C) Expression of receptor mRNA was assessed by SYBR Green RTQPCR using specific primers. The means and standard deviations from 3 independent RNA extractions are shown. Statistical analysis by Student's t-test; *P < 0.05 compared to vector control
Examination of TRAIL receptor messenger RNA (mRNA) expression demonstrated that TRAIL-R1 and TRAIL-R2 are transcriptionally up-regulated at least 5-fold in cells expressing Hsp70 compared to vector control cells (Fig 3C). Expression of TRAIL-R3 and TRAIL-R4 mRNA was similar to that of TRAIL-R1 and TRAIL-R2 mRNA in vector control cells and was unchanged by expression of Hsp70. Expression of receptor mRNA was no different in clone 18 cells compared to vector control cells. In addition, expression of the internal control, vimentin, did not vary between the 4 cell lines (data not shown). Thus, Hsp70 is inducing TRAIL sensitivity in CCRF-CEM cells by up-regulation of TRAIL-receptor expression at the transcriptional level.
p53 protein is up-regulated in CCRF-CEM cells but not SW480 cells expressing Hsp70
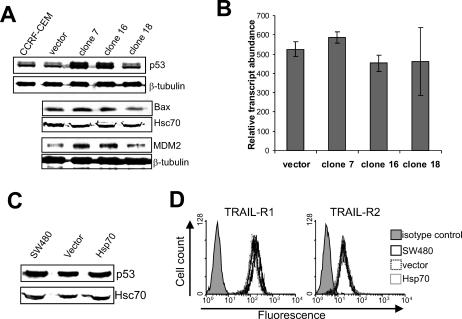
Expression of TRAIL receptors is p53-inducible (Sheikh et al 1998; Meng et al 2000; Wu et al 2000; Guan et al 2001). Whereas CCRF-CEM cells have mutated p53 (Cheng and Haas 1990), this mutation retains some transactivation activity (Park et al 1994). Significantly, this mutation is known to produce a p53 conformational change that can form a complex with Hsp70 (Gannon et al 1990). The levels of p53 in CCRF-CEM cells were examined by Western blotting as a potential explanation for the increase in TRAIL-R1 and TRAIL-R2 expression (Fig 4A). Two specific bands were detected with the p53 antibody, the higher band possibly representing phosphorylated forms of the protein. Expression of p53 and the phosphorylated protein was increased in cells expressing Hsp70. Expression of 2 known p53-inducible genes, Bax and MDM2, was also moderately up-regulated in cells expressing Hsp70, indicating transactivation activity of p53 in these cells (Fig 4A). Measurement of p53 mRNA levels (Fig 4B) revealed no differences (P > 0.05) between cells expressing Hsp70 and vector control cells, indicating that Hsp70 is up-regulating p53 by a posttranslational event, possibly through protein stabilization.
Fig 4.
p53 protein is up-regulated in CCRF-CEM but not SW480 cells expressing Hsp70. (A) Expression of p53 and the p53-regulated genes Bax and MDM2 in parental, vector control, and Hsp70-expressing cells was assessed by Western blotting. (B) Expression of p53 mRNA was assessed by SYBR Green RTQPCR, and relative expression was determined by comparison with vimentin. (C) The effect of Hsp70 on expression of p53 in SW480 cells was evaluated by Western blotting. (D) Expression of TRAIL-R1 and TRAIL-R2 on the plasma membrane of SW480 cells was analyzed by flow cytometry using specific antibodies
Expression of p53, TRAIL-R1, and TRAIL-R2 was also examined in SW480 cells. Expression of p53 in SW480 cells was unaltered by Hsp70 (Fig 4C). In contrast to CCRF-CEM cells, parental SW480 and vector control cells expressed high levels of both TRAIL-R1 and TRAIL-R2 on the cell surface and their expression was not altered by expression of Hsp70 (Fig 4D). TRAIL-R3 and TRAIL-R4 were not expressed in these cells (data not shown), possibly explaining why parental SW480 cells are sensitive to TRAIL, whereas CCRF-CEM cells are not. These results suggest that Hsp70 is unable to up-regulate TRAIL-R1 or TRAIL-R2 through regulation of p53 in these cells. SW480 cells contain mutated p53 (Nigro et al 1989; Rodrigues et al 1990), which again has been shown to retain transactivational activity (Park et al 1994). Significantly, the particular p53 mutations found in SW480 are known to produce a conformation nearly identical to that of wild-type p53, which is not bound by Hsp70 (Gannon et al 1990; Zambetti and Levine 1993).
Increased levels of p53 in the presence of Hsp70 promotes binding to the TRAIL-R2 gene
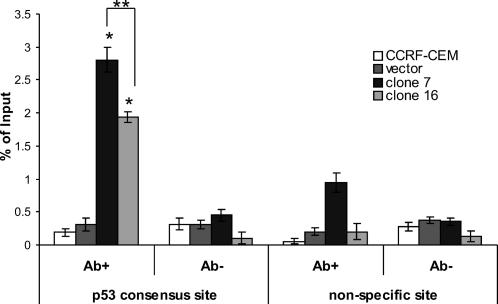
The gene encoding TRAIL-R2 contains a p53 binding site that resides in the first intron (Takimoto and El-Deiry 2000; Yoshida et al 2001). p53 is able to bind to this site and drive expression of reporter constructs (Takimoto et al 2000). By ChIP assay, we demonstrated a 4- to 6-fold increase in the level of p53 binding to the TRAIL-R2 gene in Hsp70-expressing cells compared to that of vector control and parental cells (Fig 5). Binding of p53 was significantly higher in clone 7 compared to clone 16 (P < 0.05), possibly reflecting the higher expression of Hsp70 in clone 7 (Clemons et al 2005). There was a small increase in the level of the nonspecific site amplicon detected in clone 7 cells, but not clone 16 cells. This was possibly due to the immunoprecipitation of DNA fragments that were large enough to include the nonspecific site, which is located approximately 900 bp upstream of the p53 binding sequence. The level of the p53-specific amplicon detected in clone 7 cells was still over 2-fold greater than the level of the nonspecific amplicon. Thus, the ChIP assay demonstrated that there is an increase in the binding of p53 to the TRAIL-R2 gene in Hsp70-expressing CCRF-CEM cells.
Fig 5.
Enhanced p53 protein in the presence of elevated Hsp70 increases binding to the TRAIL-R2 gene. Chromatin from fixed and sonicated cell lysates from untreated CCRF-CEM, vector control, and Hsp70-expressing cells was immunoprecipitated with a human p53-specific antibody. Immunoprecipitated DNA was analyzed by SYBR Green RTQPCR performed in triplicate using primers specific to the p53 binding region or to a nonspecific site 900 bp upstream of the consensus site. The experiment was repeated twice with similar results each time. Statistical analysis by Student's t-test; *P < 0.05 compared to vector control; **P < 0.05 clone 7 vs clone 16
DISCUSSION
The ability of Hsp70 to inhibit stress-induced apoptosis is widely accepted; however, there are few reports exploring the consequences of Hsp70 expression on apoptosis induced through the extrinsic pathway. In addition, the mechanism by which Hps72 exerts its inhibitory function is controversial, with reports that it can act both upstream and downstream of the mitochondria to inhibit apoptosis (Jaattela et al 1998; Beere et al 2000; Mosser et al 2000; Saleh et al 2000; Gotoh et al 2004; Steel et al 2004; Guo et al 2005; Stankiewicz et al 2005). We reported recently that, like Bcl-2, Hsp70 could inhibit Fas-mediated apoptosis upstream of the mitochondria in type II cells (Clemons et al 2005). Here we investigated the role of Hsp70 in TRAIL-induced apoptosis in both type I and type II cells. In the type I cell line, SW480, neither Hsp70 nor Bcl-2 influenced TRAIL-induced apoptosis, indicating that these cells are also type I in response to TRAIL and confirming that Hsp70 cannot modulate the extrinsic pathway. This is in agreement with another study using SW480 cells, which showed that in a thermotolerant state where Hsp70 is induced, TRAIL-induced apoptosis was not inhibited (Ozoren et al 2002).
The type II cell line used here, CCRF-CEM, is intrinsically resistant to TRAIL, but unexpectedly, these cells were sensitized by stable expression of Hsp70. Further investigation revealed that TRAIL-R1 and TRAIL-R2 were up-regulated on the cell surface and at the transcriptional level in cells expressing Hsp70. These effects correlated with increased expression of p53 protein in the absence of changes in mRNA expression and binding of p53 to a consensus sequence in the TRAIL-R2 promoter. Interestingly, expression of Fas, another p53 target gene, was unchanged on the cell surface in cells expressing Hsp70 (Clemons et al 2005).
Expression of Hsp70 in CCRF-CEM up-regulated p53 protein but did not alter mRNA levels, indicating that Hsp70 is acting at a posttranslational level to increase p53. This could be achieved by altered phosphorylation, acetylation, or localization of the protein, all of which can modulate the interaction between p53 and MDM2 and thus p53 stability and activity (Xu 2003). Alternatively, Hsp70 may be binding directly to p53 to stabilize the protein and inhibit protein turnover. CCRF-CEM contain compound heterozygous p53 mutations at codons 175 and 248 (Cheng et al 1990) but generate a protein that retains some transactivation activity (Park et al 1994). Both these mutations are in the DNA-binding domain and are known as hot-spot mutants, because they are among the most common p53 mutations in human cancer (Hollstein et al 1994). The codon 248 mutant is known as a contact site mutant because this site directly contacts DNA in wild-type protein. Mutant p53 at codon 175 forms a conformation that complexes with Hsp70, whereas p53 with mutation at codon 248 does not (Gannon et al 1990; Zambetti et al 1993). Thus, it is likely that Hsp70 is interacting with and stabilizing the codon 175 mutant p53 only. Presumably, this mutant p53 retains transactivation activity because the expression of MDM2 and Bax are also up-regulated in these cells.
In contrast, expression of Hsp70 did not affect p53 levels in SW480 cells, nor did it alter TRAIL receptor expression on the cell surface. The p53 of SW480 cells has double homozygous mutations at codons 273 (also a contact site mutation) and 309 (Nigro et al 1989; Rodrigues et al 1990). Mutation of codon 273 is frequently found in colorectal carcinoma (Hollstein et al 1991; Levine 1992) and displays many characteristics of wild-type p53, reflecting the fact that this mutation does not alter the wild-type p53 conformation (Bartek et al 1990; Gannon et al 1990). Importantly, neither codon 273 mutants nor wild-type p53 have been demonstrated to bind Hsp70 (Gannon et al 1990; Zambetti et al 1993; Zylicz et al 2001), therefore providing a likely explanation for the inability of Hsp70 to alter p53 levels and hence TRAIL-receptor expression in SW480 cells.
Increased levels of p53 protein in the presence of Hsp70 in CCRF-CEM cells promotes p53 transactivation activity as demonstrated by increased expression of the p53-target genes Bax, MDM2, TRAIL-R1, and TRAIL-R2. The TRAIL-R2 gene contains an intronic p53 binding site through which p53 can induce transcription (Takimoto et al 2000; Yoshida et al 2001). By ChIP, we showed that expression of Hsp70 resulted in increased binding of p53 to the TRAIL-R2 gene. We propose that this promotes TRAIL-R2 gene transcription and sensitizes CCRF-CEM cells to TRAIL following increased expression of the protein on the cell surface. Although a p53 binding site has not been described for the TRAIL-R1 gene, there is evidence that it is a DNA damage–inducible, p53 regulated gene (Guan et al 2001). Thus, it is likely that expression of Hsp70 is also up-regulating TRAIL-R1 through p53. Whereas expression of TRAIL-R1 and TRAIL-R2 mRNA was similar in the 2 Hsp70-expressing clones, expression of TRAIL-R1 on the cell surface was higher. This is possibly due to differences in posttranscriptional regulation as reported recently by Zhang et al (2004), or simply to differences in antibody affinity. TRAIL-R3 and TRAIL-R4 have also been shown to be p53-inducible (Sheikh et al 1999; Meng et al 2000), and like the TRAIL-R2 gene, the TRAIL-R3 gene contains an intronic p53 binding site (Ruiz de Almodovar et al 2004). However, we did not see an increase in the expression of either TRAIL-R3 or TRAIL-R4 following expression of Hsp70, indicating that other regulatory mechanisms are involved in the transcription of these genes.
Higher expression of Hsp70 in clone 7 compared to clone 16 cells (Clemons et al 2005) correlated with higher levels of TRAIL-induced apoptosis (Fig 1B,C) and increased binding of p53 to the TRAIL-R2 gene (Fig 5). In contrast, expression of p53, TRAIL-R1, and TRAIL-R2 were not significantly different between the 2 clones (Figs 3 and 4A), indicating that there are other mechanisms involved in producing the higher levels of TRAIL-induced apoptosis in clone 7 cells. It is tempting to speculate that Hsp70 may have further roles in modulating TRAIL-induced apoptosis, such as increasing stability of the receptors at the surface during ligand binding, leading to enhanced apoptotic signaling prior to receptor turnover, or through assisting receptor trafficking to the cell membrane. Either of these possibilities could explain the increased level of TRAIL-induced apoptosis in clone 7 compared to clone 16 cells in the absence of differences in the basal expression of the receptors on the cell surface. The absence of an increase in TRAIL-R2 transcription despite increased binding of p53 to the gene in clone 7 cells compared to clone 16 cells could be related to other transcription factors playing a limiting role.
An increase in cellular p53 is insufficient to increase its transcriptional activity; this requires conformational changes that result from further covalent modifications at the C-terminal end of the protein (Giaccia and Kastan 1998; Meek 1999). Interestingly, such conformational changes can also be achieved by the interaction of some antibodies and peptides with the C terminus (Selivanova et al 1998). In addition, numerous proteins bind to p53 and can modify its stability as well as its ability to activate transcription (Prives and Hall 1999). In CCRF-CEM cells, induction of p53 by ionizing radiation is insufficient for transactivation of the TRAIL-R2 gene (our unpublished observations). Instead, transactivation appears to require the assistance of Hsp70.
In contrast to our data, expression of Hsp70 in HL60 cells resulted in protection from TRAIL-induced apoptosis, suggesting that this cell line is also type II (Guo et al 2005). HL60 cells lack p53, and thus, presumably in this context Hsp70 is unable to up-regulate TRAIL-R1 or -R2 expression and instead is free to act in the intrinsic pathway to inhibit apoptosis induced by TRAIL. Also, thermotolerance, which among other events, involves an up-regulation of Hsp70, was found to inhibit TRAIL-induced apoptosis in the type II cell line, HCT116 (Ozoren et al 2002). In addition, constitutive overexpression of Hsp70 in HCT116 cells does not alter either TRAIL receptor or p53 expression (our unpublished observations). HCT116 cells contain wild-type p53; thus Hsp70 would be unlikely to regulate the transactivation activity of p53 and hence TRAIL-receptor expression in these cells.
Our findings have implications for the use of TRAIL as a cancer therapy. Hsp70 is expressed at high levels in a number of high-grade tumors, particularly those of the breast, compared to low-grade tumors and normal surrounding tissue (Ciocca et al 1993; Chant et al 1995; Lazaris et al 1995, 1997; Ralhan and Kaur 1995; Athanassiadou et al 1998). In addition, p53 is commonly mutated in cancer (Hussain and Harris 1998), which may result in the formation of nonnative conformations that are recognized and complexed by Hsp70, thereby stabilizing the protein and promoting its transactivation activity if the mutant p53 retains this function. Thus, tumors with a combination of high Hsp70 expression and mutated, Hsp70 binding p53 may be good candidates for therapy with TRAIL.
Acknowledgments
This work was supported by National Institutes of Health/National Cancer Institute Grant CA81421 (R.L.A.). We thank Drs. Andreas Strasser, David Huang, Bill Welch, Peter Krammer, Yuri Lazebnik, and Clayton Hunt for the kind provision of reagents, and Dr. Bedrich Eckhardt for help with figures. We also thank Genentech, Inc, and Amgen, Inc, for provision of recombinant human TRAIL and anti-human TRAIL receptor antibodies, respectively. Results using Amgen, Inc, antibodies are released with permission (Amgen reference number 200110248/201543).
REFERENCES
- Almasan A, Ashkenazi A. Apo2L/TRAIL: apoptosis signaling, biology, and potential for cancer therapy. Cytokine Growth Factor Rev. 2003;14:337–348. doi: 10.1016/s1359-6101(03)00029-7.1359-6101(2003)014[0337:TASBAP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Arizono Y, Yoshikawa H, Naganuma H, Hamada Y, Nakajima Y, Tasaka K. A mechanism of resistance to TRAIL/Apo2L-induced apoptosis of newly established glioma cell line and sensitisation to TRAIL by genotoxic agents. Br J Cancer. 2003;88:298–306. doi: 10.1038/sj.bjc.6600666.0007-0920(2003)088[0298:AMORTA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002;2:420–430. doi: 10.1038/nrc821.1474-175X(2002)002[0420:TDADRO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Pai RC, and Fong S. et al. 1999 Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 104:155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanassiadou P, Petrakakou E, Sakelariou V, Zerva C, Liossi A, Michalas S, Athanassiades P. Expression of p53, bcl-2 and heat shock protein (hsp72) in malignant and benign ovarian tumours. Eur J Cancer Prev. 1998;7:225–231. doi: 10.1097/00008469-199806000-00007.0959-8278(1998)007[0225:EOPBAH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bartek J, Iggo R, Gannon J, Lane DP. Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene. 1990;5:893–899.0950-9232(1990)005[0893:GAIAOM]2.0.CO;2 [PubMed] [Google Scholar]
- Beere HM, Wolf BB, and Cain K. et al. 2000 Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat Cell Biol. 2:469–475. [DOI] [PubMed] [Google Scholar]
- Buzzard KA, Giaccia AJ, Killender M, Anderson RL. Heat shock protein 72 modulates pathways of stress-induced apoptosis. J Biol Chem. 1998;273:17147–17153. doi: 10.1074/jbc.273.27.17147.0021-9258(1998)273[17147:HSPMPO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Chant ID, Rose PE, Morris AG. Analysis of heat-shock protein expression in myeloid leukaemia cells by flow cytometry. Br J Haematol. 1995;90:163–168. doi: 10.1111/j.1365-2141.1995.tb03395.x.0007-1048(1995)090[0163:AOHPEI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Cheng J, Haas M. Frequent mutations in the p53 tumor suppressor gene in human leukemia T-cell lines. Mol Cell Biol. 1990;10:5502–5509. doi: 10.1128/mcb.10.10.5502.0270-7306(1990)010[5502:FMITPT]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnaiyan AM, Prasad U, and Shankar S. et al. 2000 Combined effect of tumor necrosis factor-related apoptosis-inducing ligand and ionizing radiation in breast cancer therapy. Proc Natl Acad Sci USA. 97:1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciocca DR, Clark GM, Tandon AK, Fuqua SA, Welch WJ, McGuire WL. Heat shock protein hsp70 in patients with axillary lymph node-negative breast cancer: prognostic implications. J Natl Cancer Inst. 1993;85:570–574. doi: 10.1093/jnci/85.7.570.0027-8874(1993)085[0570:HSPHIP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Clemons NJ, Buzzard K, Steel R, Anderson RL. Hsp72 inhibits Fas-mediated apoptosis upstream of the mitochondria in type II cells. J Biol Chem. 2005;280:9005–9012. doi: 10.1074/jbc.M414165200.0021-9258(2005)280[9005:HIFAUO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Creagh EM, Sheehan D, Cotter TG. Heat shock proteins—modulators of apoptosis in tumour cells. Leukemia. 2000;14:1161–1173. doi: 10.1038/sj.leu.2401841.0887-6924(2000)014[1161:HSPOAI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Eskes R, Desagher S, Antonsson B, Martinou JC. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol Cell Biol. 2000;20:929–935. doi: 10.1128/mcb.20.3.929-935.2000.0270-7306(2000)020[0929:BITOAI]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth EE, Thilly WG, Penman BW, Liber HL, Rand WM. Quantitative assay for mutation in diploid human lymphoblasts using microtiter plates. Anal Biochem. 1981;110:1–8. doi: 10.1016/0003-2697(81)90103-2.0003-2697(1981)110[0001:QAFMID]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gabai VL, Meriin AB, Mosser DD, Caron AW, Rits S, Shifrin VI, Sherman MY. Hsp70 prevents activation of stress kinases. A novel pathway of cellular thermotolerance. J Biol Chem. 1997;272:18033–18037. doi: 10.1074/jbc.272.29.18033.0021-9258(1997)272[18033:HPAOSK]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gannon JV, Greaves R, Iggo R, Lane DP. Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J. 1990;9:1595–1602. doi: 10.1002/j.1460-2075.1990.tb08279.x.1460-2075(1990)009[1595:AMIPPA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973.0890-9369(1998)012[2973:TCOPME]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gliniak B, Le T. Tumor necrosis factor-related apoptosis-inducing ligand's antitumor activity in vivo is enhanced by the chemotherapeutic agent CPT-11. Cancer Res. 1999;59:6153–6158.0008-5472(1999)059[6153:TNFALA]2.0.CO;2 [PubMed] [Google Scholar]
- Gotoh T, Terada K, Oyadomari S, Mori M. hsp70-DnaJ chaperone pair prevents nitric oxide- and CHOP-induced apoptosis by inhibiting translocation of Bax to mitochondria. Cell Death Differ. 2004;11:390–402. doi: 10.1038/sj.cdd.4401369.1350-9047(2004)011[0390:HCPPNO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7.1042-2196(2002)001[0019:AMOLAD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Griffith TS, Rauch CT, and Smolak PJ. et al. 1999 Functional analysis of TRAIL receptors using monoclonal antibodies. J Immunol. 162:2597–2605. [PubMed] [Google Scholar]
- Guan B, Yue P, Clayman GL, Sun SY. Evidence that the death receptor DR4 is a DNA damage-inducible, p53-regulated gene. J Cell Physiol. 2001;188:98–105. doi: 10.1002/jcp.1101.0021-9541(2001)188[0098:ETTDRD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Guo F, Sigua C, and Bali P. et al. 2005 Mechanistic role of heat shock protein 70 in Bcr-Abl-mediated resistance to apoptosis in human acute leukemia cells. Blood. 105:1246–1255. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9.0092-8674(2000)100[0057:THOC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hollstein M, Rice K, and Greenblatt MS. et al. 1994 Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840.0193-4511(1991)253[0049:PMIHC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Huang DC, Hahne M, and Schroeter M. et al. 1999 Activation of Fas by FasL induces apoptosis by a mechanism that cannot be blocked by Bcl-2 or Bcl-xL. Proc Natl Acad Sci USA. 96:14871–14876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DC, Tschopp J, Strasser A. Bcl-2 does not inhibit cell death induced by the physiological Fas ligand: implications for the existence of type I and type II cells. Cell Death Differ. 2000;7:754–755. doi: 10.1038/sj.cdd.4400683.1350-9047(2000)007[0754:BDNICD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hussain SP, Harris CC. Molecular epidemiology of human cancer: contribution of mutation spectra studies of tumor suppressor genes. Cancer Res. 1998;58:4023–4037.0008-5472(1998)058[4023:MEOHCC]2.0.CO;2 [PubMed] [Google Scholar]
- Jaattela M. Escaping cell death: survival proteins in cancer. Exp Cell Res. 1999;248:30–43. doi: 10.1006/excr.1999.4455.0014-4827(1999)248[0030:ECDSPI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Jaattela M, Wissing D, Bauer PA, Li GC. Major heat shock protein hsp70 protects tumor cells from tumor necrosis factor cytotoxicity. EMBO J. 1992;11:3507–3512. doi: 10.1002/j.1460-2075.1992.tb05433.x.1460-2075(1992)011[3507:MHSPHP]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaattela M, Wissing D, Kokholm K, Kallunki T, Egeblad M. Hsp70 exerts its anti-apoptotic function downstream of caspase-3-like proteases. EMBO J. 1998;17:6124–6134. doi: 10.1093/emboj/17.21.6124.1460-2075(1998)017[6124:HEIAFD]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst. 2000;92:1564–1572. doi: 10.1093/jnci/92.19.1564.0027-8874(2000)092[1564:ROTHSR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kelley SK, Harris LA, Xie D, Deforge L, Totpal K, Bussiere J, Fox JA. Preclinical studies to predict the disposition of Apo2L/ tumor necrosis factor-related apoptosis-inducing ligand in humans: characterization of in vivo efficacy, pharmacokinetics, and safety. J Pharmacol Exp Ther. 2001;299:31–38.0022-3565(2001)299[0031:PSTPTD]2.0.CO;2 [PubMed] [Google Scholar]
- Kischkel F, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer P, Peter M. Cytotoxicity-dependent APO-1 (Fas/ CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x.1460-2075(1995)014[5579:CACPFA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel FC, Lawrence DA, and Tinel A. et al. 2001 Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem. 276:46639–46646. [DOI] [PubMed] [Google Scholar]
- Krammer PH. CD95(APO-1/Fas)-mediated apoptosis: live and let die. Adv Immunol. 1999;71:163–210. doi: 10.1016/s0065-2776(08)60402-2.0065-2776(1999)071[0163:FALALD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lawrence D, Shahrokh Z, and Marsters S. et al. 2001 Differential hepatocyte toxicity of recombinant Apo2L/TRAIL versions. Nat Med. 7:383–385. [DOI] [PubMed] [Google Scholar]
- Lazaris A, Chatzigianni EB, Panoussopoulos D, Tzimas GN, Davaris PS, Golematis B. Proliferating cell nuclear antigen and heat shock protein 70 immunolocalization in invasive ductal breast cancer not otherwise specified. Breast Cancer Res Treat. 1997;43:43–51. doi: 10.1023/a:1005706110275.0167-6806(1997)043[0043:PCNAAH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lazaris AC, Theodoropoulos GE, Davaris PS, Panoussopoulos D, Nakopoulou L, Kittas C, Golematis BC. Heat shock protein 70 and HLA-DR molecules tissue expression. Prognostic implications in colorectal cancer. Dis Colon Rectum. 1995;38:739–745. doi: 10.1007/BF02048033.0012-3706(1995)038[0739:HSPAHM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Levine AJ. The p53 tumor-suppressor gene. N Engl J Med. 1992;326:1350–1352. doi: 10.1056/NEJM199205143262008.1533-4406(1992)326[1350:TPTG]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1.0092-8674(1998)094[0491:COBBCM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1.0092-8674(1997)091[0479:CCADFO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443.0066-4154(1986)055[1151:THR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5.0092-8674(1998)094[0481:BABIPM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Medema JP, Scaffidi C, Kischkel FC, Shevchenko A, Mann M, Krammer PH, Peter ME. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC) EMBO J. 1997;16:2794–2804. doi: 10.1093/emboj/16.10.2794.1460-2075(1997)016[2794:FIABAW]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW. Mechanisms of switching on p53: a role for covalent modification? Oncogene. 1999;18:7666–7675. doi: 10.1038/sj.onc.1202951.0950-9232(1999)018[7666:MOSOPA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Meng RD, McDonald III ER, Sheikh MS, Fornace Jr AJ, El-Deiry WS. The TRAIL decoy receptor TRUNDD (DcR2, TRAIL-R4) is induced by adenovirus-p53 overexpression and can delay TRAIL-, p53-, and KILLER/DR5-dependent colon cancer apoptosis. Mol Ther. 2000;1:130–144. doi: 10.1006/mthe.2000.0025.1525-0024(2000)001[0130:TTDRTD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Mosser DD, Caron AW, Bourget L, Denis-Larose C, Massie B. Role of the human heat shock protein hsp70 in protection against stress-induced apoptosis. Mol Cell Biol. 1997;17:5317–5327. doi: 10.1128/mcb.17.9.5317.0270-7306(1997)017[5317:ROTHHS]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DD, Caron AW, Bourget L, Meriin AB, Sherman MY, Morimoto RI, Massie B. The chaperone function of hsp70 is required for protection against stress-induced apoptosis. Mol Cell Biol. 2000;20:7146–7159. doi: 10.1128/mcb.20.19.7146-7159.2000.0270-7306(2000)020[7146:TCFOHI]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DD, Morimoto RI. Molecular chaperones and the stress of oncogenesis. Oncogene. 2004;23:2907–2918. doi: 10.1038/sj.onc.1207529.0950-9232(2004)023[2907:MCATSO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nagane M, Pan G, Weddle JJ, Dixit VM, Cavenee WK, Huang HJ. Increased death receptor 5 expression by chemotherapeutic agents in human gliomas causes synergistic cytotoxicity with tumor necrosis factor-related apoptosis-inducing ligand in vitro and in vivo. Cancer Res. 2000;60:847–853.0008-5472(2000)060[0847:IDREBC]2.0.CO;2 [PubMed] [Google Scholar]
- Nigro JM, Baker SJ, and Preisinger AC. et al. 1989 Mutations in the p53 gene occur in diverse human tumour types. Nature. 342:705–708. [DOI] [PubMed] [Google Scholar]
- Ozoren N, El-Deiry W. Heat shock protects HCT116 and H460 cells from TRAIL-induced apoptosis. Exp Cell Res. 2002;281:175–181. doi: 10.1006/excr.2002.5660.0014-4827(2002)281[0175:HSPHAH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Park DJ, Nakamura H, Chumakov AM, Said JW, Miller CW, Chen DL, Koeffler HP. Transactivational and DNA binding abilities of endogenous p53 in p53 mutant cell lines. Oncogene. 1994;9:1899–1906.0950-9232(1994)009[1899:TADBAO]2.0.CO;2 [PubMed] [Google Scholar]
- Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687.0021-9258(1996)271[12687:IOABAL]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3.0022-3417(1999)187[0112:TPP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ralhan R, Kaur J. Differential expression of Mr 70,000 heat shock protein in normal, premalignant, and malignant human uterine cervix. Clin Cancer Res. 1995;1:1217–1222.1078-0432(1995)001[1217:DEOMHS]2.0.CO;2 [PubMed] [Google Scholar]
- Rodrigues NR, Rowan A, Smith ME, Kerr IB, Bodmer WF, Gannon JV, Lane DP. p53 mutations in colorectal cancer. Proc Natl Acad Sci USA. 1990;87:7555–7559. doi: 10.1073/pnas.87.19.7555.1091-6490(1990)087[7555:PMICC]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de Almodovar C, Ruiz-Ruiz C, Rodriguez A, Ortiz-Ferron G, Redondo JM, Lopez-Rivas A. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) decoy receptor TRAIL-R3 is up-regulated by p53 in breast tumor cells through a mechanism involving an intronic p53-binding site. J Biol Chem. 2004;279:4093–4101. doi: 10.1074/jbc.M311243200.0021-9258(2004)279[4093:TNFALT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Saleh A, Srinivasula SM, Balkir L, Robbins PD, Alnemri ES. Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat Cell Biol. 2000;2:476–483. doi: 10.1038/35019510.1465-7392(2000)002[0476:NROTAA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Samali A, Cotter TG. Heat shock proteins increase resistance to apoptosis. Exp Cell Res. 1996;223:163–170. doi: 10.1006/excr.1996.0070.0014-4827(1996)223[0163:HSPIRT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Scaffidi C, Fulda S, and Srinivasan A. et al. 1998 Two CD95 (APO-1/ Fas) signaling pathways. EMBO J. 17:1675–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz I, Walczak H, Krammer PH, Peter ME. Differences between CD95 type I and II cells detected with the CD95 ligand. Cell Death Differ. 1999;6:821–822. doi: 10.1038/sj.cdd.4400569.1350-9047(1999)006[0821:DBCTIA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Selivanova G, Kawasaki T, Ryabchenko L, Wiman KG. Reactivation of mutant p53: a new strategy for cancer therapy. Semin Cancer Biol. 1998;8:369–378. doi: 10.1006/scbi.1998.0099.1044-579X(1998)008[0369:ROMPAN]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sheikh MS, Burns TF, and Huang Y. et al. 1998 p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor-α. Cancer Res. 58:1593–1598. [PubMed] [Google Scholar]
- Sheikh MS, Huang Y, and Fernandez-Salas EA. et al. 1999 The antiapoptotic decoy receptor TRID/TRAIL-R3 is a p53-regulated DNA damage-inducible gene that is overexpressed in primary tumors of the gastrointestinal tract. Oncogene. 18:4153–4159. [DOI] [PubMed] [Google Scholar]
- Stankiewicz AR, Lachapelle G, Foo CP, Radicioni SM, Mosser DD. Hsp70 inhibits heat-induced apoptosis upstream of mitochondria by preventing Bax translocation. J Biol Chem. 2005;280:38729–38739. doi: 10.1074/jbc.M509497200.0021-9258(2005)280[38729:HIHAUO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Steel R, Doherty JP, Buzzard K, Clemons N, Hawkins CJ, Anderson RL. Hsp72 inhibits apoptosis upstream of the mitochondria and not through interactions with Apaf-1. J Biol Chem. 2004;279:51490–51499. doi: 10.1074/jbc.M401314200.0021-9258(2004)279[51490:HIAUOT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Subjeck JR, Sciandra JJ, Johnson RJ. Heat shock proteins and thermotolerance; a comparison of induction kinetics. Br J Radiol. 1982;55:579–584. doi: 10.1259/0007-1285-55-656-579.0007-1285(1982)055[0579:HSPAAC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Takimoto R, El-Deiry WS. Wild-type p53 transactivates the KILLER/DR5 gene through an intronic sequence-specific DNA-binding site. Oncogene. 2000;19:1735–1743. doi: 10.1038/sj.onc.1203489.0950-9232(2000)019[1735:WPTTDG]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312.0193-4511(1998)281[1312:CEW]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4.0092-8674(1999)096[0245:CDID]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Walczak H, Miller RE, and Ariail K. et al. 1999 Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 5:157–163. [DOI] [PubMed] [Google Scholar]
- Wei MC, Lindsten T, and Mootha VK. et al. 2000 tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 14:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- Wei MC, Zong WX, and Cheng EH. et al. 2001 Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 292:727–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley SR, Schooley K, and Smolak PJ. et al. 1995 Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 3:673–682. [DOI] [PubMed] [Google Scholar]
- Wu GS, Kim K, el-Deiry WS. Killer/dr5, a novel DNA-damage inducible death receptor gene, links the p53-tumor suppressor to caspase activation and apoptotic death. Adv Exp Med Biol. 2000;465:143–151. doi: 10.1007/0-306-46817-4_13.0065-2598(2000)465[0143:DANDID]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Xu Y. Regulation of p53 responses by post-translational modifications. Cell Death Differ. 2003;10:400–403. doi: 10.1038/sj.cdd.4401182.1350-9047(2003)010[0400:ROPRBP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Maeda A, Tani N, Sakai T. Promoter structure and transcription initiation sites of the human death receptor 5/ TRAIL-R2 gene. FEBS Lett. 2001;507:381–385. doi: 10.1016/s0014-5793(01)02947-7.0014-5793(2001)507[0381:PSATIS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zambetti GP, Levine AJ. A comparison of the biological activities of wild-type and mutant p53. FASEB J. 1993;7:855–865. doi: 10.1096/fasebj.7.10.8344485.0892-6638(1993)007[0855:ACOTBA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zhang XY, Zhang XD, Borrow JM, Nguyen T, Hersey P. Translational control of tumor necrosis factor-related apoptosis-inducing ligand death receptor expression in melanoma cells. J Biol Chem. 2004;279:10606–10614. doi: 10.1074/jbc.M308211200.0021-9258(2004)279[10606:TCOTNF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2.0092-8674(1997)090[0405:AAHPHT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zylicz M, King FW, Wawrzynow A. Hsp70 interactions with the p53 tumour suppressor protein. EMBO J. 2001;20:4634–4638. doi: 10.1093/emboj/20.17.4634.1460-2075(2001)020[4634:HIWTPT]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]