Abstract
Myosin VI supports movement toward the (−) end of actin filaments, despite sharing extensive sequence and structural homology with (+)-end-directed myosins. A class-specific stretch of amino acids inserted between the converter domain and the lever arm was proposed to provide the structural basis of directionality reversal. Indeed, the unique insert mediates a 120° redirection of the lever arm in a crystal structure of the presumed poststroke conformation of myosin VI [Ménétrey J, Bahloul A, Wells AL, Yengo CM, Morris CA, Sweeney HL, Houdusse A (2005) Nature 435:779–785]. However, this redirection alone is insufficient to account for the large (−)-end-directed stroke of a monomeric myosin VI construct. The underlying motion of the myosin VI converter domain must therefore differ substantially from the power stroke of (+)-end-directed myosins. To experimentally map out the motion of the converter domain and lever arm, we have generated a series of truncated myosin VI constructs and characterized the size and direction of the power stroke for each construct using dual-labeled gliding filament assays and optical trapping. Motors truncated near the end of the converter domain generate (+)-end-directed motion, whereas longer constructs move toward the (−) end. Our results directly demonstrate that the unique insert is required for directionality reversal, ruling out a large class of models in which the converter domain moves toward the (−) end. We suggest that the lever arm rotates ≈180° between pre- and poststroke conformations.
Keywords: actin, molecular motor, pointed end, swinging cross-bridge
The basic actomyosin motor has been embellished, altered, and reused many times through the evolution of diverse members of the myosin superfamily (1). Class VI myosins are highly specialized (−)-end-directed motors involved in a growing list of functions in animal cells, including endocytosis, cell migration, and maintenance of stereociliar membrane tension (2). Initial biophysical characterization (3–5) of myosin VI raised two important questions concerning the structural basis of its unique motor properties. First, how does the motor achieve reverse directionality? And second, how does dimeric myosin VI take long steps along actin, matching the stride of myosin V without the apparent benefit of a long lever arm?
A Backward-Moving Myosin
Myosin VI attracted attention as a candidate for a (−)-end-directed motor after the identification of a class-specific insert following the converter domain. In the swinging cross-bridge model of actomyosin function, the converter domain transmits motion from the myosin head to the C-terminal lever arm, generating a directed power stroke. Wells et al. (3) hypothesized that a structure inserted between the converter domain and the lever arm could redirect the stroke and lead to backward movement. The predicted reverse directionality of myosin VI was experimentally confirmed (3), but the precise mechanism of this remarkable adaptation remained unclear, including whether the unique insert was necessary and sufficient for stroke reversal.
Tsiavaliaris et al. (6) used protein engineering to show that the proposed mechanism of power stroke reversal was feasible. They inserted a four-helix bundle after the converter domain of a (+)-end-directed myosin, redirecting the lever arm by 180° and generating an artificial (−)-end-directed motor. Their results clearly demonstrate that an insertion between the converter domain and the lever arm can be sufficient for directionality reversal in a myosin, but leave open the question of whether myosin VI takes advantage of this available mechanism.
Homma et al. (7) questioned whether the unique insert was necessary for directionality reversal. They characterized a collection of chimeric motors in which portions of myosin VI were fused to portions of myosin V, with surprising results. One chimera contained the motor domain of myosin VI (including the converter but not the subsequent unique insert) fused to the lever arm of myosin V. This construct was reported to generate (−)-end-directed motion, suggesting that the determinants of directionality might be located within the motor domain.
A Long Stride and a Long Stroke
A new puzzle arose when the stepping behavior of dimeric myosin VI was characterized by using single molecule techniques (4, 5). Myosin VI, like myosin V, walks processively along actin by using a long stride that approximately matches the ≈36-nm pseudorepeat of the actin filament. Myosin V achieves its long stride with the aid of a long lever arm containing six IQ motifs bound to light chains. Myosin VI has only a single IQ motif, preceded by the unique insert and followed by a predicted coiled coil region.
Partial explanations for the long stride of myosin VI came from further examination of the regions flanking the IQ motif. The unique insert was found to bind calmodulin despite containing no recognizable light chain binding motif, suggesting that this insert may help to extend the lever arm (8, 9). The predicted coiled coil region also has surprising properties: the first ≈80 aa of this region (dubbed the “proximal tail domain”) do not appear to form a stable coiled coil. Instead, the proximal tail forms an unknown flexible structure that contributes to the length of the stride (10). A model emerged in which the unique insert and IQ domain form a lever arm that strokes to provide an initial bias for the myosin VI stride, followed by a diffusive component facilitated by the proximal tail domain.
A diffusive search may help to lengthen the stride of the dimer, but the monomeric myosin VI power stroke also appears to be surprisingly large. The insert and IQ domain could be expected to form a lever arm ≈10 nm long, matching the apparent lever arm seen in EM studies (3). The power stroke of monomeric myosin VI has been measured at 12–18 nm by different groups (10, 11) using different constructs, implying that the angle swept out by the myosin VI power stroke may be substantially larger than for other characterized myosins. Lister et al. (11) proposed that myosin VI swings its lever arm 138° between the pre- and poststroke conformations.
Myosin VI Must Have an Unusual Power Stroke
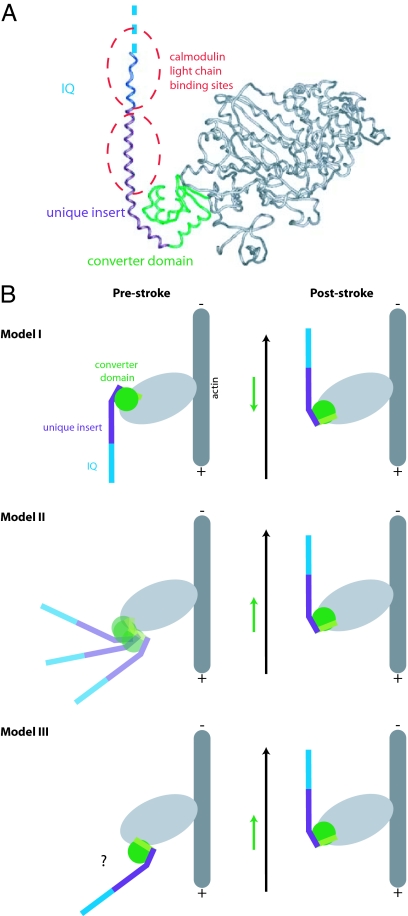
The crystal structure solved by Ménétrey et al. (12) provided key insights into the structural basis of the myosin VI power stroke. The structure shows the poststroke conformation of the myosin VI catalytic head, unique insert, and IQ domain (Fig. 1A). The unique insert clearly mediates a major redirection of the lever arm (≈120°) loosely reminiscent of the artificial (−)-end-directed myosin designed by Tsiavaliaris et al. (6). Moreover, the distal unique insert and IQ domain, together with their associated calmodulin light chains, form an extended lever arm that points nearly parallel to the actin filament in the poststroke state.
Fig. 1.
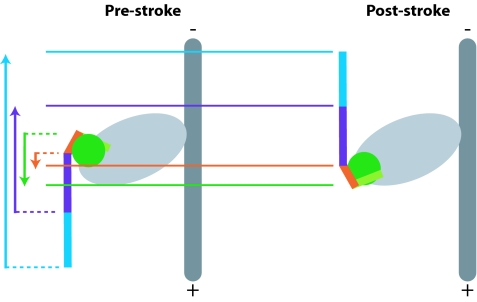
The pre- and poststroke conformations of myosin VI. (A) Poststroke x-ray crystal structure (12). A class-specific insertion (purple) emerges from the final helix of the converter domain (green) and serves to redirect the lever arm toward the (−) end of the actin filament. The IQ domain (blue) is not completely resolved in the structure and is followed by a flexible domain of unknown structure that may extend the lever arm (see Discussion). Two calmodulin light chains (not shown) bind to the extended helix formed by the IQ domain and the distal portion of the unique insert at the locations indicated by the dashed ovals. (B) Speculative models for the myosin VI power stroke [models I and II adapted from Ménétrey et al. (12)]. Light chains are omitted for clarity. The final helix of the converter domain is indicated in light green. Black arrows indicate the net motion of the tip of the lever arm, and green arrows indicate the net motion of the converter domain. In model I, the converter domain moves toward the (+) end of the actin filament while the redirected lever arm moves toward the (−) end. To explain the large stroke size, the converter domain must be rotated in the prestroke state relative to its position in known prestroke structures of (+)-end-directed myosins. In model II, the power stroke consists of a disorder-to-order transition in which the average position of the converter domain moves toward the (−) end. Another class of models (model III) involves a rigid motion of the converter domain toward the (−) end. Other possibilities (not shown) might include a disruption of the interface between the unique insert and the converter domain.
Is the structure of the redirected lever arm sufficient to explain the size and direction of the power stroke? Employing the reasonable assumption that the insert and IQ domain remain rigidly attached to the converter domain, Ménétrey et al. (12) modeled the unknown prestroke conformation of myosin VI by rotating the converter to match the prestroke conformation of (+)-end-directed myosins. This model predicted a power stroke that was directed toward the (−) end but far smaller (≈2.5 nm) than the measured myosin VI stroke size. The authors concluded that myosin VI does not simply perform a standard myosin power stroke that is redirected and amplified by C-terminal structures. Rather, the force-generating conformational change of the myosin VI catalytic domain must diverge significantly from other myosins.
Speculative Models
Ménétrey et al. (12) proposed two models for the divergent power stroke of myosin VI (Fig. 1B). In model I, the converter position in the prestroke state differs from that of (+)-end-directed myosins by an ≈90° rotation that serves to point the lever arm directly toward the (+) end of the actin filament. In this model, the converter domain moves toward the (+) end as in other myosins, but the tip of the lever arm swings through a wide arc to generate a large (−)-end-directed stroke. This model would entail dramatic structural changes in the catalytic domain, and therefore the authors favored a second model. In model II, the concept of a rigid power stroke was abandoned entirely. In this model, the converter domain becomes uncoupled from the myosin head in the prestroke state, and the “stroke” consists of a disorder-to-order transition reminiscent of the docking of the neck linker in kinesin (13). Other models not covered by Ménétrey et al. (12) are possible; for example, the chimeric myosin data of Ikebe and coworkers (7) could be used to argue for a rigid prestroke state in which the converter lies in a novel orientation positioned far toward the (+) end. We will refer to this possibility as model III. To distinguish between these and other models, we have mapped out the myosin VI power stroke by characterizing the motions of a series of deletion constructs.
Dissecting Myosin Power Strokes by Modifying the Lever Arm
Myosin constructs with truncated or extended lever arms have been powerful tools for testing the basic swinging cross-bridge model and for detecting subtle differences between the power strokes of different myosin classes. Uyeda et al. (14) measured the velocity of gliding filaments powered by Dictyostelium myosin II constructs in which the lever arm was shortened by deleting one or both light chain binding motifs, or else extended by the addition of a third motif. Velocity was found to be linearly dependent on the length of the putative lever arm, arguing for a power stroke based on a rigid rotation mechanism. These studies were extended through the use of optical traps (which allowed stroke sizes to be measured directly) and artificial lever arms, further bolstering the swinging cross-bridge model (15, 16). Linear plots of stroke size versus truncation position contain information about both the angle of the lever arm swing (reflected in the slope of the plot) and the position of the fulcrum (reflected in the x-intercept). It was found that the angle swept out by the power stroke can differ between myosins. A fairly large swing of ≈90° for myosin I (versus typical values of ≈60° for myosin II) is supported by both a truncation series analysis of myosin 1d (17) and the prestroke crystal structure of Dictyostelium myosin 1e (18).
We reasoned that truncated constructs might similarly be used to characterize the power stroke of myosin VI. As with other myosins, the stroke size is expected to vary with truncation position, reflecting the angle of rotation and fulcrum position if the power stroke consists of a lever arm swing. Additionally, depending on the power stroke model, the direction of the measured stroke may be expected to depend on truncation position. Specifically, in model I, the converter domain moves in the opposite direction from the end of the lever arm, predicting that shorter constructs should be (+)-end-directed while longer constructs are (−)-end-directed. Here, we show that this prediction of model I is confirmed, arguing strongly against both model II and model III, which predict that all truncation constructs will move toward the (−) end. The sizes and directions of the power strokes measured with truncated myosin VI constructs provide distance constraints that may ultimately lead toward a detailed three-dimensional model of the unknown myosin VI prestroke state.
Results
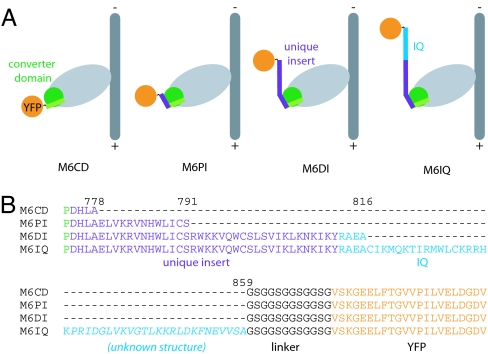
We designed four constructs to help us map out the power stroke of myosin VI (Fig. 2). Each construct consists of a truncated porcine myosin VI heavy chain fused to a flexible linker followed by YFP. Four different truncation positions were chosen at natural structural boundaries spanning the putative lever arm: M6CD is truncated after the final helix of the converter domain; M6PI is truncated after the proximal unique insert, before the first light chain binding region; M6DI is truncated after the distal unique insert, between the first and second light chain binding regions; and M6IQ is truncated well beyond the end of the second light chain binding region (IQ motif). The C-terminal YFP serves as a handle for oriented attachment of motors to antibody-coated surfaces (4).
Fig. 2.
A series of constructs for dissecting the myosin VI power stroke. The myosin VI gene was truncated at one of four positions and fused to a flexible linker followed by YFP and a C-terminal Flag peptide. YFP serves as a handle for oriented attachment to antibody-coated surfaces. (A) Diagrams of fusion constructs bound to actin in the poststroke conformation. (B) Partial amino acid sequence for the four constructs showing the junctions between the truncated myosin, flexible linker, and YFP. Truncations are all designed to occur slightly C-terminal to domain boundaries, to favor the formation of fully folded domains.
Truncation Alters the Velocity and Directionality of Myosin VI.
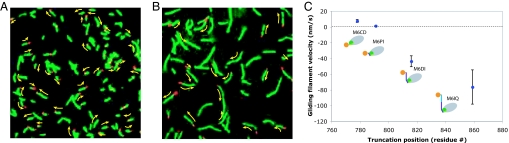
To measure the velocity and direction of movement driven by our myosin VI constructs, we observed the gliding motion of polarity-marked actin filaments (19) on surfaces coated with motors bound to anti-YFP (Fig. 3). The longest construct (M6IQ) showed (−)-end-directed movement at a velocity of 80 ± 20 nm/s, in good agreement with previous studies of monomeric myosin VI (3, 11). M6DI, which lacks the IQ domain, showed slower (−)-end-directed motion [Fig. 3 and supporting information (SI) Movie 1], in qualitative agreement with the predictions of most models of myosin VI function. To distinguish between power stroke models, it was necessary to examine the shorter constructs, which lack both light chain binding domains.
Fig. 3.
Dual-labeled gliding filament assays. Motility assays were performed in chambers coated with anti-YFP. Filaments were labeled at their barbed (+) ends by using capped seeds of Cy5-actin (19) and stabilized with TMR-phalloidin. (A and B) Images from movies of gliding dual-labeled filaments. Barbed ends are visible in the red (Cy5) channel, and TMR fluorescence is shown in green. Overlaid yellow arrows indicate the direction of motion of gliding filaments. (A) M6DI supports (−)-end-directed motion. Gliding dual-labeled filaments move with their labeled ends leading. (B) M6CD supports (+)-end-directed motion. (C) Plot of gliding filament velocity versus truncation position (residue #) for the four constructs. Velocities were measured at 2 mM ATP. Positive values are shown for constructs exhibiting (+)-end-directed motion. Error bars are ±SD (M6CD, n = 60; M6PI, n = 20; M6DI, n = 76; M6IQ, n = 55). See also SI Movies 1 and 2.
M6CD and M6PI both supported (+)-end-directed motion (Fig. 3 and SI Movie 2), showing that regions outside of the catalytic head are required for (−)-end-directed motility. The directionality of these constructs is consistent with model I and rules out model II and model III. The very slow [but clearly (+)-end-directed] motility of M6PI suggests that the truncation point at the end of the proximal unique insert lies near the axis of rotation of the lever arm.
Optical Trapping Provides a Direct Measurement of Stroke Sizes.
We have interpreted differences in gliding velocities between the constructs as reflective of differences in the size of the power stroke. However, other factors can in principle contribute to gliding velocities. In an idealized model, the gliding velocity V is given by V = d/τ, where d is the length of the power stroke and τ is the average time that the myosin spends attached to the actin filament during each cycle. If the kinetics of the myosin head were perturbed in our truncation constructs, then differences in gliding velocity could be due to changes in τ rather than d. Additional factors, such as subpopulations of inactive myosins on the surface, may also contribute to altered gliding velocities.
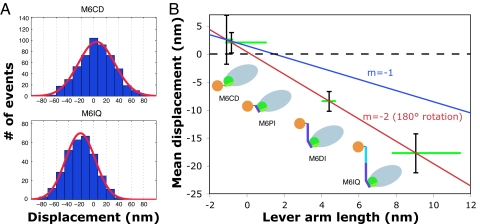
The suspended filament optical trapping assay (20) provides direct measurements of both d and τ. In this assay, myosin motors are deposited at very low densities on raised micrometer-scale platforms. An actin filament is held between two optically trapped beads and suspended over a platform until isolated interactions are observed between the filament and a single myosin molecule. Attachment of actin to surface-bound myosin can be detected by correlation of the Brownian motion of the trapped beads (21). The attachment time Δt and positional displacement of the beads from equilibrium Δx are then measured for each interaction event. In the absence of a power stroke, the distribution of Δx is a Gaussian whose width reflects thermal fluctuations of the bead-actin assembly. The power stroke that follows each attachment event has the effect of shifting the distribution of Δx by the amount d (Fig. 4A).
Fig. 4.
Single-molecule measurements of myosin VI construct power strokes. The suspended filament assay (20) was used to characterize interactions between actin filaments and individual oriented myosin VI fusion molecules attached to surface platforms via antibodies to YFP. (A) Examples of histograms of bead displacements during actomyosin attachment events. Each histogram is taken from a single motor platform. Mean displacements are offset from zero because of the myosin power stroke, which differs in magnitude for the different fusion constructs. (B) Mean displacement as a function of lever arm length (in nanometers). Displacements shown are the average results from two separate platforms (M6CD, M6PI, and M6DI) or three platforms (M6IQ), after correction for series compliance. Because the structures of the truncation constructs are not precisely known, each lever arm length is represented by a green bar spanning reasonable values for the construct. Maximum possible lever arm lengths were estimated from a projection along the axis of the helix that extends through the IQ domain, measured on the poststroke crystal structure from the break in the helix at V784 to the final myosin VI residue included in each construct. For M6IQ, unknown structure beyond the residues included in the crystal structure was presumed to be α-helical to provide an upper bound. Minimum lever arm lengths were estimated by assuming peeling of C-terminal structure back to domain boundaries (see Fig. 2). Data are consistent with a line of slope 2 (red line) representing a 180° rotation of the lever arm. Slopes larger than 2 are impossible with a lever arm rotation mechanism, and slopes much smaller than 2 are inconsistent with our data.
Suspended filament assays show that the attachment kinetics of all four constructs are unperturbed at 20 μM ATP. A 2-fold difference in the attachment kinetics at saturating ATP suggests that M6CD and M6PI have slightly altered ADP release rates relative to M6DI, M6IQ, and full-length myosin VI (Table 1).
Table 1.
Attachment kinetics and stroke sizes
| Construct | τ (20 μM ATP), s | τ (2 mM ATP), s | V, nm/s | Vτ, nm | 〈Δx〉, nm | 〈Δx〉 corrected, nm |
|---|---|---|---|---|---|---|
| M6CD | 1.48 ± 0.05 | 0.62 ± 0.03 | 7 ± 2 | 5 ± 1 | 2 ± 3 | 2 ± 4 |
| M6PI | 1.32 ± 0.05 | 0.58 ± 0.02 | 1.2 ± 0.6 | 0.7 ± 0.4 | 2 ± 1 | 2 ± 2 |
| M6DI | 1.39 ± 0.04 | 0.27 ± 0.01 | −44 ± 7 | −12 ± 2 | −7 ± 2 | −9 ± 2 |
| M6IQ | 1.58 ± 0.08 | 0.28 ± 0.01 | −80 ± 20 | −21 ± 6 | −14 ± 3 | −18 ± 3 |
Mean attachment times of M6IQ and M6DI were measured at room temperature in the suspended filament optical trapping assay. At low [ATP], kinetics are in reasonable agreement with previous kinetic studies of myosin VI (11, 27, 28). M6CD and M6PI have slightly altered kinetics at saturating [ATP], suggesting that truncation may have a small effect on ADP release but not on ATP binding. Mean ± SE of τ is given for N > 200 events in all cases. Mean displacements 〈Δ x〉 measured in the optical trapping assay provide an estimate of the stroke sizes of the constructs. Displacements are shown before and after correction for series compliance. Correction factors were estimated using the method of correlated thermal diffusion (21). For comparison, gliding velocities V for each construct are also tabulated (mean ± SD; see Fig. 3) along with the idealized projected stroke size d = Vτ obtained by multiplying the gliding velocity by the attachment time.
The measured stroke sizes for the four constructs follow the trend we previously observed in the gliding velocities, as expected (Table 1 and Fig. 4B). The stroke size of the longest construct (M6IQ) is close to the largest stroke size previously reported (11) for a monomeric myosin VI motor (18 nm). When the stroke sizes are plotted as a function of lever arm length, an approximately linear relationship is seen, as with other myosins (Fig. 4B). This linearity is consistent with a rigidly rotating lever arm model. A plot of stroke size versus truncation position may be used to estimate the angle swept out by the lever arm. Our data are well described by a line of slope 2 on this plot, suggesting an ≈180° rotation of the converter and lever arm.
Discussion
We have measured the magnitudes and directions of power strokes produced by truncated myosin VI constructs (Fig. 5). Our results are consistent with one of the two models proposed by Ménétrey et al. (12) and imply that myosin VI has evolved a very large (≈180°) converter rotation to amplify its (−)-end-directed stroke. A large converter rotation for myosin VI was previously proposed by Lister et al. (11) purely on the basis of the monomeric stroke size. The structural basis of this unusual converter rotation remains to be elucidated but may depend on the cluster of class-specific mutations in the SH1 cavity identified by Ménétrey et al. (12).
Fig. 5.
The power stroke of myosin VI. Our results are consistent with model I proposed by Ménétrey et al. (see Fig. 1), and we suggest that the lever arm rotates ≈180° between the prestroke and poststroke conformations. Poststroke positions (solid lines) of M6CD (green) and M6PI (orange) truncation positions are further toward the (+) end than their proposed prestroke positions (dashed lines). The reverse is true for the truncation positions of M6DI (purple) and M6IQ (blue). We cannot rule out an alternative model in which the interface between the unique insert and the converter domain is disrupted or altered in the prestroke state.
A large stroke size is probably essential to the processive stepping mechanism of dimeric myosin VI, which moves along actin with ≈36-nm strides, matching the pseudorepeat of the actin helix (4, 5). Unlike myosin V, myosin VI has not used multiple IQ repeats to achieve a large stroke. Instead, it apparently employs an unusually large converter rotation together with a noncanonical light-chain binding domain (the distal unique insert) and a single IQ motif ending at P835. The lever arm may in fact be slightly longer because of an unknown structure formed by residues 835–859; this possibility is represented by the long green bar representing M6IQ in Fig. 4B. (Because the structures of the truncation constructs are not directly known, the green bars in Fig. 4B represent a range of reasonable values of the lever arm length for each construct, determined as described in the legend to Fig. 4.) Electrospray ionization mass spectrometry showed that residues beyond G839 contribute to the stability of the interaction between the IQ domain and calmodulin (8, 9). We speculate that the region following G839 may fold back to form a structure that interacts with the calmodulin light chain and extends the lever arm.
Our laboratory has previously reported a stroke size of ≈12 nm using different monomeric myosin VI constructs. However, the M6IQ stroke size measured here is in close agreement with the monomeric working stroke (∼18 nm) reported by Lister et al. (11). Given the relatively weak affinity of the IQ domain for calmodulin (particularly in the M6S1 construct truncated at G839), our previous shorter stroke size measurements may have resulted from incomplete saturation of the IQ with calmodulin, leading to an unstructured lever arm following the unique insert.
In our truncation constructs, the unique insert was required for (−)-end-directed motion. Ikebe and coworkers (7) have previously reported (−)-end-directed motion for a chimera between the myosin VI head (ending at G761, well before the unique insert) and the myosin V lever arm. This result remains puzzling, but it is possible that the chimera forms an unexpected structure; for example, the myosin V lever arm could pack across the converter domain in a way that mimics the redirection mediated by the unique insert. It will be informative to investigate the properties of a wider range of chimeras, perhaps including myosin VI constructs with artificial lever arms (15).
Mechanical adaptations in molecular motors may occur via modifications to peripheral structures or to the catalytic core. Successful attempts at artificially reengineering myosin stroke size and directionality have so far relied on the comparatively simple strategy of adding or replacing mechanical elements C-terminal to the core (22). Nature has apparently used a more complex strategy with myosin VI: the large reverse stroke depends on class-specific adaptations inside the core (producing a large converter rotation unlike that of other myosins) in addition to the unique C-terminal insertion. The structural basis of variability in the conformational changes of molecular motor cores remains a challenging subject for future study.
Methods
Proteins.
DNA constructs encoding truncated porcine myosin VI (Fig. 2) followed by (GSG)4, eYFP, and a C-terminal Flag tag (GDYKDDDDK) were cloned into pBiEX-1 (Novagen) and expressed by transfection of plasmid DNA into 15-ml cultures of SF9 cells according to the recommendations of the InsectDirect system (Novagen) except that Escort IV cationic lipids (Sigma) were used as the transfection reagent. Cells were harvested after 60–72 h at 28°C, and proteins were affinity-purified (23) by using anti-Flag resin (Sigma). After elution from resin with 0.8 mg/ml Flag peptide (DYKDDDDK), myosin fusion proteins (SI Fig. 6) were used for functional assays without further purification. Gelsolin was provided by Matt Footer (Stanford University).
In Vitro Motility.
All functional assays were performed at room temperature in assay buffer containing 25 mM imidazole·HCl (pH 7.4), 25 mM KCl, 1 mM EGTA, 10 mM DTT, 4 mM MgCl2, 5 μM CaM, and reagents for oxygen scavenging (0.4% glucose, 0.2 mg/ml glucose oxidase, and 36 μg/ml catalase) and ATP regeneration (1 mM phosphocreatine and 100 μg/ml creatine phosphokinase). Dual-labeled actin filaments were prepared according to the method of Soldati and coworkers (19). Briefly, actin was labeled by using Cy5-maleimide (Amersham), polymerized in the presence of gelsolin, and stabilized with phalloidin to form barbed-end capped seeds. Seeds were then extended with excess unlabeled actin and in the presence of TMR-phalloidin. Gliding filament assays were performed as described (4, 24) on surfaces coated with mouse monoclonal anti-GFP (Chemicon) and observed by using either a Zeiss upright epifluorescence microscope with exchangeable filters or a custom-built microscope with dual-view optics (25). Images were recorded on an EMCCD camera (Andor) and analyzed by using ImageJ software (National Institutes of Health). Dual-labeled filaments yielded >95% consistency in scoring directionality.
Optical Trapping.
Suspended filament assays on antibody-coated platforms were performed as described (10, 26) by using a custom-built, dual-beam optical trap. Actin dumbbells were held taut (at typical tensions of 1.5–2.5 pN) to minimize attenuation of stroke size due to series compliance. Phalloidin (10 μM) was maintained in the assay buffer to prevent breakage of actin filaments under tension. Trap stiffnesses were in the range 0.006–0.012 pN/nm. Motor dilutions were chosen to give conditions in which ≈10% of tested platforms yielded actomyosin binding events. Binding eventswere identified in the data traces by eye using the drop in position variance and bead–bead correlation (21).
Supplementary Material
Acknowledgments
We thank R. Rock and R. Cooke for critical readings of the manuscript; S. Sutton for technical assistance; and A. Dunn, B. Spink, and L. Churchman for helpful discussions. This work was supported by National Institutes of Health Grant GM33289 (to J.A.S.). Z.B. is a Helen Hay Whitney Postdoctoral Fellow. D.A. was supported by an American Heart Association Predoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610144104/DC1.
References
- 1.Thompson RF, Langford GM. Anat Rec. 2002;268:276–289. doi: 10.1002/ar.10160. [DOI] [PubMed] [Google Scholar]
- 2.Buss F, Spudich G, Kendrick-Jones J. Annu Rev Cell Dev Biol. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- 3.Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 4.Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL. Proc Natl Acad Sci USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishikawa S, Homma K, Komori Y, Iwaki M, Wazawa T, Hikikoshi Iwane A, Saito J, Ikebe R, Katayama E, Yanagida T, Ikebe M. Biochem Biophys Res Commun. 2002;290:311–317. doi: 10.1006/bbrc.2001.6142. [DOI] [PubMed] [Google Scholar]
- 6.Tsiavaliaris G, Fujita-Becker S, Manstein DJ. Nature. 2004;427:558–561. doi: 10.1038/nature02303. [DOI] [PubMed] [Google Scholar]
- 7.Homma K, Yoshimura M, Saito J, Ikebe R, Ikebe M. Nature. 2001;412:831–834. doi: 10.1038/35090597. [DOI] [PubMed] [Google Scholar]
- 8.Chevreux G, Potier N, Van Dorsselaer A, Bahloul A, Houdusse A, Wells A, Sweeney HL. J Am Soc Mass Spectrom. 2005;16:1367–1376. doi: 10.1016/j.jasms.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 9.Bahloul A, Chevreux G, Wells AL, Martin D, Nolt J, Yang Z, Chen LQ, Potier N, Van Dorsselaer A, Rosenfeld S, et al. Proc Natl Acad Sci USA. 2004;101:4787–4792. doi: 10.1073/pnas.0306892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rock RS, Ramamurthy B, Dunn AR, Beccafico S, Rami BR, Morris C, Spink BJ, Franzini-Armstrong C, Spudich JA, Sweeney HL. Mol Cell. 2005;17:603–609. doi: 10.1016/j.molcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Lister I, Schmitz S, Walker M, Trinick J, Buss F, Veigel C, Kendrick-Jones J. EMBO J. 2004;23:1729–1738. doi: 10.1038/sj.emboj.7600180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ménétrey J, Bahloul A, Wells AL, Yengo CM, Morris CA, Sweeney HL, Houdusse A. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, et al. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 14.Uyeda TQ, Abramson PD, Spudich JA. Proc Natl Acad Sci USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruff C, Furch M, Brenner B, Manstein DJ, Meyhofer E. Nat Struct Biol. 2001;8:226–229. doi: 10.1038/84962. [DOI] [PubMed] [Google Scholar]
- 16.Warshaw DM, Guilford WH, Freyzon Y, Krementsova E, Palmiter KA, Tyska MJ, Baker JE, Trybus KM. J Biol Chem. 2000;275:37167–37172. doi: 10.1074/jbc.M006438200. [DOI] [PubMed] [Google Scholar]
- 17.Kollmar M, Durrwang U, Kliche W, Manstein DJ, Kull FJ. EMBO J. 2002;21:2517–2525. doi: 10.1093/emboj/21.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler D, Ruff C, Meyhofer E, Bahler M. J Cell Biol. 2003;161:237–241. doi: 10.1083/jcb.200212039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herm-Gotz A, Weiss S, Stratmann R, Fujita-Becker S, Ruff C, Meyhofer E, Soldati T, Manstein DJ, Geeves MA, Soldati D. EMBO J. 2002;21:2149–2158. doi: 10.1093/emboj/21.9.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finer JT, Simmons RM, Spudich JA. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 21.Mehta AD, Finer JT, Spudich JA. Proc Natl Acad Sci USA. 1997;94:7927–7931. doi: 10.1073/pnas.94.15.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manstein DJ. Philos Trans R Soc London B. 2004;359:1907–1912. doi: 10.1098/rstb.2004.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Cruz EM, Ostap EM, Sweeney HL. J Biol Chem. 2001;276:32373–32381. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- 24.Rock RS, Rief M, Mehta AD, Spudich JA. Methods. 2000;22:373–381. doi: 10.1006/meth.2000.1089. [DOI] [PubMed] [Google Scholar]
- 25.Okten Z, Churchman LS, Rock RS, Spudich JA. Nat Struct Mol Biol. 2004;11:884–887. doi: 10.1038/nsmb815. [DOI] [PubMed] [Google Scholar]
- 26.Rice SE, Purcell TJ, Spudich JA. Methods Enzymol. 2003;361:112–133. doi: 10.1016/s0076-6879(03)61008-6. [DOI] [PubMed] [Google Scholar]
- 27.Robblee JP, Olivares AO, de la Cruz EM. J Biol Chem. 2004;279:38608–38617. doi: 10.1074/jbc.M403504200. [DOI] [PubMed] [Google Scholar]
- 28.Altman D, Sweeney HL, Spudich JA. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.