Abstract
Considering the importance of its potential implications for human health, agricultural productivity, and ecosystem stability, surprisingly little is known regarding the composition or dynamics of the atmosphere's microbial inhabitants. Using a custom high-density DNA microarray, we detected and monitored bacterial populations in two U.S. cities over 17 weeks. These urban aerosols contained at least 1,800 diverse bacterial types, a richness approaching that of some soil bacterial communities. We also reveal the consistent presence of bacterial families with pathogenic members including environmental relatives of select agents of bioterrorism significance. Finally, using multivariate regression techniques, we demonstrate that temporal and meteorological influences can be stronger factors than location in shaping the biological composition of the air we breathe.
Keywords: 16S rRNA, biosurveillance, aerobiology, microarray, climate change
Low levels of moisture and nutrients combined with high levels of UV radiation make the earth's atmosphere an extreme environment for microbial life. Little is known regarding the atmospheric microbial composition and how it varies by location or meteorological conditions. Plant canopies for example, are known to be significant sources of bacterial aerosols with upward flux of bacteria positively impacted by temperature and wind speed (1). Aerosols created at the surface of aquatic systems are known to concentrate and carry bacteria through the liquid–air interface (2, 3). The relationship between environmental conditions and bacterial aerial dispersal indicates that climate change could potentially alter the microbial composition of downwind areas, resulting in increased health risk from pathogens or allergenic components of unclassified environmental bacteria. For instance, the last decade has seen a dramatic increase in the amount of desertification and a concomitant increase in upper atmospheric particulates (4). In sub-Saharan regions of Africa, dust storms have been associated with regional outbreaks of meningococcal meningitis caused by the bacterium Neisseria meningitidis (5). Since the 1970s, El Nino weather events have coincided with increased flux of African Dust across the Atlantic (4) that, in turn, has been linked to coral reef disease (6) and increased exacerbations of pediatric asthma (7) in the Caribbean. Therefore, as particles from dust storms shield bacterial and fungal passengers from the inactivating effects of UV exposure, global transport of dust will have more far-reaching affects than impaired visibility.
The consequences of natural environmental variation such as meteorological shifts, combined with anthropogenic influences such as land use changes, may alter atmospheric microbial composition. To monitor the effects of climate change on aerosol microbial composition, it first is necessary to establish baselines that acknowledge the current microbial components and how they fluctuate naturally. However, the potential heterogeneity, both spatial and temporal, in species composition coupled with low microbial biomass ensures this is not a facile task.
Natural shifts in bacterial composition also have implications for atmospheric pathogen monitoring systems, such as the Department of Homeland Security effort to monitor major U.S. cites for intentional release of biowarfare agents (www.ostp.gov/html/10-20-03%20jhm%20BioSecurity%202003.pdf). Many such pathogens and other closely related bacteria with undefined pathogenicity already are endemic to the locations that are being monitored (8) and so may interfere with detection networks (9), but little is known regarding the frequency or variability of their occurrence. Most aerobiology studies to date (e.g., refs. 10–12), have used culture-based methods for determining microbial composition. Although some studies recently have applied culture-independent techniques (e.g., refs. 13 and 14), little is known of what constitutes the breadth of diversity of “typical” organisms in the atmosphere (as opposed to those capable of growth in laboratory media) and what influences their composition. To address these methodological limitations and to augment our view of aerosol microbial diversity and dynamics, we have designed a microarray (PhyloChip) for the comprehensive identification of both bacterial and archaeal organisms. We target the variation in the 16S rRNA gene, possessed by all prokaryotes, to capture the broad range of microbial diversity that may be present in the atmosphere. This tool allows bacteria and archaea to be identified and monitored in any type of sample without the need for microbial cultivation.
The two greatest obstacles to designing a 16S rRNA gene-based microarray to identify individual organisms in a complex environmental mixture are natural sequence diversity and potential cross-hybridization. Sequence diversity is an issue as we sample new and distinctive environments such as the atmosphere. There may be many undocumented organisms with 16S rRNA gene sequences that are similar, but not identical, to the sequences that were used for array design. Microarrays based on single sequence-specific hybridizations (single probes) may be ineffective in detecting such environmental sequences with one or several polymorphisms. To overcome this obstacle, we have designed a minimum of 11 different, short oligonucleotide probes for each taxonomic grouping, allowing for the failure of one or more probes. On the other hand, nonspecific cross-hybridization is an issue when an abundant 16S rRNA gene shares sufficient sequence similarity to nontargeted probes, such that a weak but detectable signal is obtained. We have found that the perfect match (PM)-mismatch (MM) probe pair approach effectively minimizes the influence of cross-hybridization. Widely used on expression arrays as a control for nonspecific binding (15), the central nucleotide is replaced with any of the three nonmatching bases so that the increased hybridization intensity signal of the PM over the paired MM indicates a sequence-specific, positive hybridization. By requiring multiple PM-MM probe pairs to have a positive interaction, we substantially increase the chance that the hybridization signal is due to a predicted target sequence.
We grouped known 16S rRNA gene sequences >600 bp into distinct taxa such that a set of at least 11 probes that were specific to the taxon could be chosen. The resulting 8,935 taxa (8,741 of which are represented on the PhyloChip), each containing ≈3% sequence divergence, represented all 121 demarcated bacterial and archaeal orders [supporting information (SI) Table 2]. For a majority of the taxa represented on the PhyloChip (5,737, 65%), probes were designed from regions of gene sequences that have been identified only within a given taxon. For 1,198 taxa (14%), no probe-level sequence could be identified that was not shared with other groups of 16S rRNA gene sequences, although the gene sequence as a whole was distinctive. For these taxonomic groupings, a set of at least 11 probes was designed to a combination of regions on the 16S rRNA gene that taken together as a whole did not exist in any other taxa. For the remaining 1,806 taxa (21%), a set of probes were selected to minimize the number of putative cross-reactive taxa. Although more than half of the probes in this group have a hybridization potential to one outside sequence, this sequence was typically from a phylogenetically similar taxon. For all three probe set groupings, the advantage of the hybridization approach is that multiple taxa can be identified simultaneously by targeting unique regions or combinations of sequence.
To assess the bacterial composition of environmental aerosols and how it changes over time and with location, we examined outdoor air collected at multiple locations in two cities, Austin and San Antonio, TX. These cities are part of the U.S. multiagency biosurveillance effort that use aerosol collectors to concentrate airborne particulate matter in search of pathogens that potentially could be indicative of a bioterrorism threat. For either city, aerosol monitors were used to draw in air and pass it through filters designed to collect submicrometer particulates for a 24-h period. The samplers were placed immediately adjacent to six Environmental Protection Agency air quality monitoring stations located throughout the urban area of each city, and the filter eluents were pooled for each day before amplification of the 16S rRNA gene products from the extracted DNA. Although PCR amplification may introduce some bias in terms of quantitative assessment of an organism's abundance due to factors such as preferential amplification (16, 17), the extremely low bacterial biomass in aerosol samples necessitate such an approach. Amplified products from 4 days within a 7-day period were pooled into a single sample representing 1 week, and 17 consecutive weekly samples beginning May 2003 were analyzed from both cities for bacterial composition.
Results and Discussion
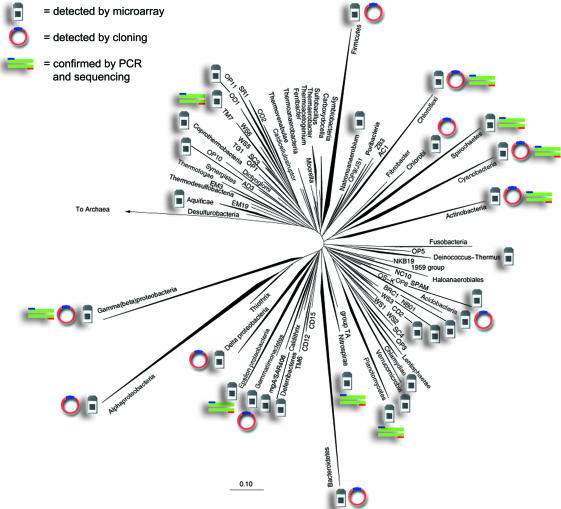
PhyloChip results for one sample, representing bacteria recovered from outdoor air at San Antonio from the week of July 14–20 (calendar week 29), 2003, were compared with clone library sequence results from the same pool of amplified 16S rRNA gene products (Fig. 1). A conservative comparison of the PhyloChip and cloning approaches was made at a taxonomic level below family and above species classification (see SI Materials and Methods), termed “subfamily” for clarity. This demonstrated that the PhyloChip correctly detected 90% of cloned subfamilies (SI Table 3) and additionally detected almost 2.5-fold more diversity at the phylum level (Fig. 1). We subsequently have confirmed many of these PhyloChip-only hits (which include known pathogenic genera), by cluster-specific PCR and sequencing (Fig. 1 and SI Table 4). The most common sequences in the air clone library (35%) were Bacilli most similar to the species Bacillus bataviensis (previously isolated from soil in a disused hay field) (18) and another Bacillus sp. associated with biodeterioration of mural paintings (19), suggesting dispersal through aerosolization. The diversity of the remaining clone sequences was quite high, with a majority of the clones representing distinctive 16S rRNA gene sequences (SI Fig. 4).
Fig. 1.
Representative phylogenetic tree showing all known bacterial phyla (and individual classes in the case of proteobacteria) annotated to show 16S rRNA gene sequences detected in an urban aerosol by both microarray and cloning. Also annotated are phyla detected by microarray only that subsequently were confirmed by targeted PCR and sequencing. The Archaea are used as an outgroup. (Scale bar: 0.1 changes per nucleotide.)
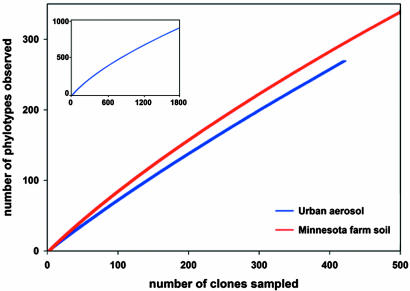
Because of the relative dearth of information regarding aerosol bacterial diversity, we compared the diversity detected in this aerosol sample by cloning with that found in a farm soil from a previous study (20). Soils are considered to be highly diverse microbial habitats with an estimate of up to 1 million distinct genomes per gram (21). Rarefaction analysis revealed a similar level of diversity (at the 16S rRNA gene biomarker level) in the aerosol and soil samples (Fig. 2). Predicted estimates of richness (Chao1 and ACE) indicated between 1,500 and 1,800 16S rRNA phylotypes (by using a 99% identity cutoff) in the aerosol sample (SI Fig. 5). However, because both ACE and Chao1 richness prediction curves were nonasymptotic, this is likely to be an underestimate because of insufficient clone sampling, a common problem when assessing environmental microbial diversity by using cloning approaches.
Fig. 2.
Rarefaction curves comparing bacterial diversity in a Minnesota farm soil (20) and the urban aerosol in this study. (Inset) Complete rarefaction curve for 1,874 sequences from the Minnesota farm soil library.
Microbial communities are characteristically dynamic, and it is expected that aerosol communities are no exception, considering the turbulent and well mixed nature of the atmosphere. Using a Latin Square type study containing mixtures of amplicons from diverse bacterial species applied to the PhyloChips in rotating concentrations (SI Table 5), we tested the ability of the PhyloChip to track 16S rRNA amplicon dynamics quantitatively. This demonstrated a strong linear relationship, spanning five orders of magnitude between PhyloChip intensities and quantities of bacterial 16S rRNA gene signatures applied to PhyloChips (SI Fig. 6). Having determined the potential of the PhyloChip for detecting changes in biomarker quantities, we analyzed intensity data for the two cities over the 17-week period of the study. We also collated a range of meteorological parameters to investigate whether local weather conditions could be correlated with the observed fluctuations in aerosol bacterial populations. Using multivariate regression tree analysis (22, 23), we examined such correlations, with tree topology and splitting parameters suggesting that sample location (in this case two geographically proximate cities) was less of a factor in explaining the variability of aerosol bacterial composition than temporal or meteorological influences (Fig. 3). The week of the year at which a sample was taken proved to be a stronger predictor of community composition with samples taken after the first three weeks in May (weeks 19–21) clustering separately from those taken before this, regardless of city sampled. Unsurprisingly, sample week was observed to correlate with weather conditions (SI Table 6). For both cities, week was positively correlated with temperature, air pressure, and visibility, whereas negatively correlated with wind speed and particulate matter. It is important to note from the composition of the PhyloChip generated tree clusters that the clone library “snapshot” taken during week 29 in San Antonio was representative of only approximately one-third of the samples collected (11 of 34 weeks clustered at this node). Therefore, caution should be used in interpreting snapshot analyses in such dynamic systems. Underlying these changes in bacterial community composition was a differential abundance of biomarkers for many spore-forming bacteria such as Actinomycetes and Firmicutes. Indeed, most of the taxa with significant correlations to weather conditions were Actinomycetes, which showed positive correlations with temperature (SI Table 7). Warmer temperatures may result in increased desiccation of soil/plant-based bacteria, leading to spore dispersal or aerosolization. Additionally, alpha-proteobacteria such as phyllosphere-inhabiting Sphingomonas spp (24). were correlated with sea-level pressure, week, and temperature (Fig. 3 and SI Table 7). These and other alpha-proteobacteria are typically oligotrophic and also may originate from freshwater and marine ecosystems (3) in addition to plant surfaces. The most significant correlation between any PhyloChip intensity pattern and an environmental/temporal variable was between the gamma-proteobacterium Pseudomonas oleovorans and week (r = 0.83, P = 2.1 × 10−5). Real-time quantitative PCR of the same genomic DNA pools used for PhyloChip PCRs demonstrated that changes in PhyloChip intensity were representative of the dynamics of this organism (SI Fig. 7).
Fig. 3.
Multivariate regression tree analysis of the interaction between aerosol bacterial dynamics (array intensity) and environmental parameters. The model explains 89.1% of variance in SI Data Set 1. Bars plotted under each cluster represent mean of normalized array intensities of phylogenetically related bacteria shown to be significantly correlated with environmental/temporal parameters.
Despite the variable nature of the aerosol bacterial population, we detected some groups of organisms in every sample over the 17-week period (summarized in Table 1, with complete details in SI Table 8). Between the two cities, more types of bacteria consistently were detected in San Antonio aerosol samples (80 subfamilies) compared with Austin (43 subfamilies), although there was significant overlap in the consistent 16S rRNA signatures between the two cities. Many of these organisms (e.g., Acidobacteria and Verrucomicrobia) are major components of the soil microbiota and may be particle-associated. Sphingomonas species also were detected consistently, psychrotolerant strains of which have been detected in dust and air samples from the Antarctic (25). Notably, other bacteria consistently detected were spore formers such as the endospore-forming Bacilli and Clostridia and the exospore-forming Actinomycetes. Cyanobacteria such as Plectonema were also frequently detected members of the aerosol community, as were plant chloroplasts (presumably from pollen). Significantly, epsilon proteobacteria were consistently detected by PhyloChip in both cities, including organisms within the families Campylobacteraceae and Helicobacteraceae, both of which contain human and animal pathogens. The exact Campylobacteraceae taxon detected by the PhyloChip contains the genus Arcobacter, whose presence we subsequently confirmed by taxon-specific PCR and sequencing (SI Table 4). This genus is known to cause bacteremia and severe gastrointestinal illnesses in humans, and together with Helicobacter (a causative agent of gastric ulcers), could be considered indicators of fecal contamination (26, 27), which is known to occur through aerosolization from wastewater treatment plants (11, 28).
Table 1.
Bacterial groups detected in all weeks during sampling period
| Phylum, class | San Antonio | Austin |
|---|---|---|
| Acidobacteria, Acidobacteria | Y | Y |
| Acidobacteria, Acidobacteria-6 | Y | N |
| Acidobacteria, Solibacteres | Y | Y |
| Actinobacteria, Actinobacteria | Y | Y |
| Actinobacteria, BD2–10 group | N | Y |
| Bacteroidetes, Sphingobacteria | Y | N |
| Chloroflexi, Anaerolineae | Y | N |
| Chloroflexi, Dehalococcoidetes | Y | N |
| Cyanobacteria, Cyanobacteria/Chloroplasts | Y | Y |
| Firmicutes, Bacilli | Y | Y |
| Firmicutes, Catabacter | Y | N |
| Firmicutes, Clostridia | Y | Y |
| Nitrospira, Nitrospira | N | Y |
| OP3, Unclassified | Y | N |
| Proteobacteria, Alphaproteobacteria | Y | Y |
| Proteobacteria, Betaproteobacteria | Y | Y |
| Proteobacteria, Gammaproteobacteria | Y | Y |
| Proteobacteria, Deltaproteobacteria | Y | N |
| Proteobacteria, Epsilonproteobacteria | Y | Y |
| TM7, TM7–3 | Y | N |
| Verrucomicrobia, Verrucomicrobiae | Y | Y |
Subfamilies detected are summarized to phylum and class level. Y, detected in each of 17 weeks sampled per city; N, not detected in every sample.
The consistent detection of signatures from potentially pathogenic bacteria led us to examine taxonomic clusters containing other pathogens (and their relatives) of public health and bioterrorism significance over the 17-week period (SI Table 9). Environmental relatives of monitored pathogens have already been implicated in multiple detection events in U.S. Homeland Security monitoring systems (www.houstontx.gov/health/NewsReleases/bacteria%20detection.htm). In fact, in response to such a detection event, a recent survey of soils in Houston was carried out to determine potential reservoirs of environmental relatives (10). This study revealed a surprising diversity of Francisella-like organisms that may have been responsible for triggering detectors in the aerosol monitoring systems. Similarly, in the aerosol samples analyzed here, we detected taxonomic clusters containing organisms closely related to Francisella in one week in Austin and two weeks in San Antonio, although the causative agent of tularemia, Francisella tularensis, was never encountered. We also consistently detected phylogenetic near-neighbors to Bacillus anthracis with the taxonomic cluster containing B. anthracis itself (also containing common soil relatives B. cereus, B. thuringiensis, and B. mycoides) being detected in one week in San Antonio. Tick-borne Rickettsia and Clostridium botulinum types C (causes illness in mammals, fish, and birds) and G (rarely illness causing) also were detected regularly, as were Burkholderia mallei and Bu. pseudomallei, which cause glanders and melioidosis respectively. Other select agents such as Yersinia pestis and Brucella spp. (melitensis, suis, and abortus) were never encountered. The frequent occurrence of environmental relatives of bacteria targeted by biosurveillance efforts in urban aerosols makes prediction of natural occurrences of endemic pathogens or their uncharacterized environmental relatives critical for the implementation of a robust biosurveillance network.
This study represents a comprehensive molecular analysis of airborne bacterial composition and dynamics. We have demonstrated that the atmosphere contains a diverse assemblage of microorganisms probably representing the amalgamation of numerous point sources. The composition of this habitat varies widely and may be subject to climatic regulation. A global-scale study of this uncharacterized ecosystem is necessary to determine baselines for bioaerosol transport patterns. Such data will enable an understanding of future anthropogenic impacts including pollution, bioterrorism, and climate change in altering the biological composition of the air we breathe.
Materials and Methods
Sample Collection and Pooling.
Air samples were collected by using an air filtration collection system under vacuum located within six Environmental Protection Agency air quality network sites in both San Antonio and Austin. Approximately 10 liters of air per minute were collected on a Celanex polyethylene terephthalate, 1.0-μm filter (Calanese, Dallas, TX). Samples were collected daily over a 24-h period. Sample filters were washed in 10 ml buffer (0.1 M sodium phosphate/10 mM EDTA, pH 7.4/0.01% Tween-20), and the suspension was stored frozen until extracted. Samples were collected from 4 May to 29 August 2003. Sample dates were divided according to a 52-week calendar year starting January 1, 2003, with each Monday-to-Sunday cycle constituting a full week. Samples from four randomly chosen days within each sample week were extracted. Each date chosen for extraction consisted of a 0.6-ml filter wash from each of the six sampling sites for that city (San Antonio or Austin) combined into a “day pool” before extraction. In total, for each week, 24 filters were sampled.
DNA Extraction and 16S rRNA Gene Amplification.
The “day pools” were centrifuged at 16,000 × g for 25 min, and the pellets were resuspended in 400 μl of 100 mM sodium phosphate buffer (pH 8). DNA extraction was performed as described in DeSantis et al. (29), but only a single bead-beating velocity and duration was used (6.5 m·s−1 for 45 s). DNA was quantified by using a PicoGreen fluorescence assay according to the manufacturer's recommended protocol (Invitrogen, Carlsbad, CA). 16S rRNA gene amplification was performed according to standard procedures as outlined in SI Materials and Methods.
PhyloChip Processing, Scanning, Probe Set Scoring, and Normalization.
The pooled PCR product was spiked with known concentrations of synthetic 16S rRNA gene fragments and non-16S rRNA gene fragments as internal standards for normalization with quantities ranging from 5.02 × 108 and 7.29 × 1010 molecules applied to the final hybridization mix (SI Table 10). Target fragmentation, biotin labeling, PhyloChip hybridization, scanning, and staining were as described by Brodie et al. (30), and background subtraction, noise calculation, and detection and quantification criteria were essentially as reported in Brodie et al. (30), with some minor exceptions. These exceptions were as follows: For a probe pair to be considered positive, the difference in intensity between the PM and MM probes must be at least 130 times the squared noise value (N). A taxon was considered present in the sample when 92% or more of its assigned probe pairs for its corresponding probe set were positive (positive fraction > = 0.92). This was determined based on empirical data from clone library analyses. Hybridization intensity (referred to as intensity) was calculated in arbitrary units for each probe set as the trimmed average (maximum and minimum values removed before averaging) of the PM minus MM intensity differences across the probe pairs in a given probe set. All intensities <1 were shifted to 1 to avoid errors in subsequent logarithmic transformations. When summarizing PhyloChip results to the subfamily, the probe set producing the highest intensity was used.
Validation of PhyloChip Detection of Airborne Bacteria by Comparison with Clone Library.
To compare the diversity of bacteria detected with PhyloChips to a known standard, one sample week was chosen for cloning and sequencing and replicate PhyloChip analysis. One large pool of SSU amplicons (96 reactions, 50 μl per reaction) from San Antonio week 29 was made. One milliliter of the pooled PCR product was gel-purified, and 768 clones were sequenced at the DOE Joint Genome Institute (Walnut Creek, CA) by standard methods. An aliquot of this same pooled PCR product also was hybridized to a PhyloChip (three replicate PhyloChips performed). Subfamilies containing a taxon scored as present in all three PhyloChip replicates were recorded. Individual cloned rRNA genes were sequenced from each terminus, assembled by using Phred and Phrap (31–33), and were required to pass quality tests of Phred 20 (base call error probability <10−2.0) to be included in the comparison. Chimeric sequences were removed after Bellerophon (34) analysis, and similarity of clones to PhyloChip taxa was calculated with DNADIST (35) measurement of homology (DNAML-F84) over 1,287 conserved columns identified by using the Lane mask (36). Sequences were assigned to a taxonomic node by using a sliding scale of similarity threshold (37). These steps are described in detail in SI Materials and Methods, and a full comparison between clone and PhyloChip analysis is available in SI Table 3.
Validation of PhyloChip-Detected Subfamilies Not Supported by the Clone Library.
Primers targeting sequences within particular taxa/subfamilies were generated by using ARB's probe design feature (38) and based on regions targeted by PhyloChip probes or were obtained from published literature (SI Table 4). Primer quality control was carried out by using Primer3 (39).
Quantitative Detection of Changes in 16S rRNA Gene Concentration in Heterogeneous Solutions.
To determine whether changes in 16S rRNA gene concentration could be detected by using the PhyloChip, various quantities of distinct rRNA gene types were hybridized to the PhyloChip in rotating combinations. We chose environmental organisms, organisms involved in bioremediation, and a pathogen of biodefense relevance. 16S rRNA genes were amplified from each of the organisms shown in SI Table 5. Then each of these nine distinct 16S rRNA gene standards was tested once in each concentration category, spanning five orders of magnitude (0 molecules, 6 × 107, 1.44 × 108, 3.46 × 108, 8.30 × 108, 1.99 × 109, 4.78 × 109, 2.75 × 1010, 6.61 × 1010, and 1.59 × 1011) with concentrations of individual 16S rRNA gene types rotating between PhyloChips such that each PhyloChip contained the same total of 16S rRNA gene molecules. This is similar to a Latin Square design, although with a 9 × 11 format matrix.
Real-Time Quantitative PCR Confirmation of PhyloChip-Observed Shifts in Taxon Abundance.
A taxon (no. 9389) consisting only of two sequences of Pseudomonas oleovorans that correlated well with environmental variables was chosen for quantitative PCR confirmation of PhyloChip-observed quantitative shifts. Primers for this taxon were designed by using the ARB (38) probe match function to determine unique priming sites based on regions detected by PhyloChip probes. These regions then were imputed into Primer3 (39) to choose optimal oligonucleotide primers for PCR. Primer quality was assessed further by using Beacon Designer v3.0 (Premier BioSoft, CA). Primers 9389F2 (CGACTACCTGGACTGACACT) and 9389R2 (CACCGGCAGTCTCCTTAGAG) were chosen to amplify a 436-bp fragment. Validation of primer specificity and reaction conditions are available in SI Materials and Methods.
Statistical Analyses.
All statistical operations were performed in the R software environment (ref. 40; www.R-project.org). For each day of aerosol sampling, 15 factors including humidity, wind, temperature, precipitation, pressure, particulate matter, and week of year were recorded from the U.S. National Climatic Data Center (www.ncdc.noaa.gov) or the Texas Natural Resource Conservation Commission (www.tceq.state.tx.us). The weekly mean, minimum, maximum, and range of values were calculated for each factor from the collected data. The changes in ln(intensity) for each PhyloChip taxon considered present in the study was tested for correlation against the environmental conditions. The resulting P values were adjusted by using the step-up false discovery rate controlling procedure (41).
Multivariate regression tree analysis (22, 23) was carried out by using the package “mvpart” within the “R” statistical programming environment. A Bray-Curtis-based distance matrix was created by using the function “gdist.” The Bray-Curtis measure of dissimilarity is generally regarded as a good measure of ecological distance when dealing with “species” abundance, or in this case, array probe-set intensity, because it allows for nonlinear responses to environmental gradients (22, 42). Large trees were calculated with splitting based on information gain and then pruned (from 13 to 10 nodes) based on 100 cross-validations to a complexity parameter of 0.025286, where cross-validation relative error had reached a plateau.
Before clone library rarefaction analysis, a distance matrix (DNAML homology) of clone sequences, was created by using an online tool at http://greengenes.lbl.gov/cgi-bin/nph-distance_matrix.cgi (43) after alignment of the sequences by using the NAST aligner (http://greengenes.lbl.gov/NAST) (44). DOTUR (45) was used to generate rarefaction curves, Chao1, and ACE richness predictions and rank-abundance curves. Nearest neighbor joining was used with 1,000 iterations for bootstrapping.
Supplementary Material
Acknowledgments
We thank Dr. Phil Hugenholtz and Dr. Paul Richardson of the Joint Genome Institute for clone library sequencing; Susannah Green Tringe for providing the soil 16S rRNA gene sequences; Sonya Murray for expert technical assistance; John Coates, Lisa Alvarez-Cohen (both of University of California, Berkeley, CA), Hoi-Ying Holman, Terry Hazen (both of Lawrence Berkeley National Laboratory), and Arthur Friedlander (U.S. Army Medical Research Institute of Infectious Diseases, Frederick, MD) for the generous gifts of bacterial cultures or DNA; and Sue Lynch, Terry Hazen, Jill Banfield, Tamas Torok, and two anonymous reviewers for helpful suggestions and comments on the manuscript. This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Berkeley National Laboratory, under Contract DE-AC02-05CH11231 and was supported in part by Department of Homeland Security Grant HSSCHQ04X00037 and the Climate Change Research Division, Biological and Environmental Research, Office of Science, U.S. Department of Energy. Computational support was provided through the Virtual Institute for Microbial Stress and Survival.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ129237–DQ129666, DQ236245–DQ236250, and DQ515230–DQ515231).
This article contains supporting information online at www.pnas.org/cgi/content/full/0608255104/DC1.
References
- 1.Lindemann J, Upper CD. Appl Environ Microbiol. 1985;50:1229–1232. doi: 10.1128/aem.50.5.1229-1232.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baylor ER, Peters V, Baylor MB. Science. 1977;197:763–764. doi: 10.1126/science.329413. [DOI] [PubMed] [Google Scholar]
- 3.Aller JY, Kuznetsova MR, Jahns CJ, Kemp PF. J Aerosol Sci. 2005;36:801–812. [Google Scholar]
- 4.Shinn EA, Griffin DW, Seba DB. Arch Environ Health. 2003;58:498–504. [PubMed] [Google Scholar]
- 5.World Health Organization. Meningococcal Meningitis Fact Sheet. Geneva: World Health Org; 2003. [Google Scholar]
- 6.Shinn EA, Smith GW, Prospero JM, Betzer P, Hayes ML, Garrison V, Barber RT. Geophys Res Lett. 2000;27:3029–3032. [Google Scholar]
- 7.Gyan K, Henry W, Lacaille S, Laloo A, Lamsee-Ebanks C, McKay S, Antoine RM, Monteil MA. Int J Biometeorol. 2005;49:371–376. doi: 10.1007/s00484-005-0257-3. [DOI] [PubMed] [Google Scholar]
- 8.Anda P, del Pozo JS, Garcia JMD, Escudero R, Pena FJG, Velasco MCL, Sellek RE, Chillaron MRJ, Serrano LPS, Navarro JFM. Emerg Infect Dis. 2001;7:575–582. doi: 10.3201/eid0707.010740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barns SM, Grow CC, Okinaka RT, Keim P, Kuske CR. Appl Environ Microbiol. 2005;71:5494–5500. doi: 10.1128/AEM.71.9.5494-5500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer H, Fuerhacker M, Zibuschka F, Schmid H, Puxbaum H. Water Res. 2002;36:3965–3970. doi: 10.1016/s0043-1354(02)00121-5. [DOI] [PubMed] [Google Scholar]
- 11.Griffin DW. Aerobiologia. 2004;20:135–140. [Google Scholar]
- 12.Lee T, Grinshpun SA, Martuzevicius D, Adhikari A, Crawford CM, Luo J, Reponen T. Indoor Air. 2006;16:37–47. doi: 10.1111/j.1600-0668.2005.00396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes KA, McCartney HA, Lachlan-Cope TA, Pearce DA. Cell Mol Biol. 2004;50:537–542. [PubMed] [Google Scholar]
- 14.Maron PA, Lejon DPH, Carvalho E, Bizet K, Lemanceau P, Ranjard L, Mougel C. Atmos Environ. 2005;39:3687–3695. [Google Scholar]
- 15.Mei R, Hubbell E, Bekiranov S, Mittmann M, Christians FC, Shen M-M, Lu G, Fang J, Liu W-M, Ryder T, et al. Proc Natl Acad Sci USA. 2003;100:11237–11242. doi: 10.1073/pnas.1534744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polz MF, Cavanaugh CM. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lueders T, Friedrich MW. Appl Environ Microbiol. 2003;69:320–326. doi: 10.1128/AEM.69.1.320-326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyrman J, Vanparys B, Logan NA, Balcaen A, Rodriguez-Diaz M, Felske A, De Vos P. Int J Syst Evol Microbiol. 2004;54:47–57. doi: 10.1099/ijs.0.02723-0. [DOI] [PubMed] [Google Scholar]
- 19.Heyrman J, Balcaen A, Rodriguez-Diaz M, Logan NA, Swings J, De Vos P. Int J Syst Evol Microbiol. 2003;53:459–463. doi: 10.1099/ijs.0.02452-0. [DOI] [PubMed] [Google Scholar]
- 20.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, et al. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 21.Gans J, Wolinsky M, Dunbar J. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 22.De'Ath G. Ecology. 2002;83:1105–1117. [Google Scholar]
- 23.Larsen DR, Speckman PL. Biometrics. 2004;60:543–549. doi: 10.1111/j.0006-341X.2004.00202.x. [DOI] [PubMed] [Google Scholar]
- 24.Kim H, Nishiyama W, Kunito T, Senoo K, Kawahara K, Murakami K, Oyaizu H. J Appl Microbiol. 1998;85:731–736. [Google Scholar]
- 25.Busse HJ, Denner EBM, Buczolits S, Salkinoja-Salonen M, Bennasar A, Kampfer P. Int J Syst Evol Microbiol. 2003;53:1253–1260. doi: 10.1099/ijs.0.02461-0. [DOI] [PubMed] [Google Scholar]
- 26.Wesley IV, Wells SJ, Harmon KM, Green A, Schroeder-Tucker L, Glover M, Siddique I. Appl Environ Microbiol. 2000;66:1994–2000. doi: 10.1128/aem.66.5.1994-2000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engberg J, On SLW, Harrington CS, Gerner-Smidt P. J Clin Microbiol. 2000;38:286–291. doi: 10.1128/jcm.38.1.286-291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes KA. Atmos Environ. 2003;37:3147–3155. [Google Scholar]
- 29.DeSantis TZ, Stone CE, Murray SR, Moberg JP, Andersen GL. FEMS Microbiol Lett. 2005;245:271–278. doi: 10.1016/j.femsle.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 30.Brodie EL, DeSantis TZ, Joyner DC, Baek S, Larsen JT, Andersen GL, Hazen TC, Richardson PM, Herman DJ, Tokunaga TK, et al. Appl Environ Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ewing B, Hillier L, Wendl MC, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 32.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 33.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 34.Huber T, Faulkner G, Hugenholtz P. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 35.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
- 36.Lane DJ. In: Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt E, Goodfellow M, editors. New York: Wiley; 1991. pp. 115–175. [Google Scholar]
- 37.Schloss PD, Handelsman J. Microbiol Mol Biol Rev. 2004;68:686–691. doi: 10.1128/MMBR.68.4.686-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozen S, Skaletsky H. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editors. Totowa, NJ: Humana; 2000. pp. 365–386. [Google Scholar]
- 40.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Found Stat Comput; 2005. [Google Scholar]
- 41.Benjamini Y, Hochberg Y. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 42.Faith DP, Minchin PR, Belbin L. Vegetatio. 1987;69:57–68. [Google Scholar]
- 43.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schloss PD, Handelsman J. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.