Abstract
Activation of peroxisome proliferator-activated receptor γ (PPARγ) by thiazolidinediones (TZDs) improves insulin resistance by increasing insulin-stimulated glucose disposal in skeletal muscle. It remains debatable whether the effect of TZDs on muscle is direct or indirect via adipose tissue. We therefore generated mice with muscle-specific PPARγ knockout (MuPPARγKO) using Cre/loxP recombination. Interestingly, MuPPARγKO mice developed excess adiposity despite reduced dietary intake. Although insulin-stimulated glucose uptake in muscle was not impaired, MuPPARγKO mice had whole-body insulin resistance with a 36% reduction (P < 0.05) in the glucose infusion rate required to maintain euglycemia during hyperinsulinemic clamp, primarily due to dramatic impairment in hepatic insulin action. When placed on a high-fat diet, MuPPARγKO mice developed hyperinsulinemia and impaired glucose homeostasis identical to controls. Simultaneous treatment with TZD ameliorated these high fat–induced defects in MuPPARγKO mice to a degree identical to controls. There was also altered expression of several lipid metabolism genes in the muscle of MuPPARγKO mice. Thus, muscle PPARγ is not required for the antidiabetic effects of TZDs, but has a hitherto unsuspected role for maintenance of normal adiposity, whole-body insulin sensitivity, and hepatic insulin action. The tissue crosstalk mediating these effects is perhaps due to altered lipid metabolism in muscle.
Introduction
Insulin resistance is a characteristic early feature of type 2 diabetes (1, 2), with reduction in insulin-stimulated glucose disposal primarily due to diminished insulin action in skeletal muscle (3, 4). Thiazolidinediones (TZDs) are pharmacologic agents that improve glucose homeostasis in type 2 diabetes by increasing insulin sensitivity, largely through improved insulin action in skeletal muscle (5–7). The antidiabetic actions of TZDs are believed to be mediated by their interaction with the nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ) (8, 9).
In both rodents and humans, adipose tissue is the major expression site for PPARγ, where it is a major regulator of adipogenesis (10). PPARγ is expressed at much lower levels in other tissues, including other major insulin target tissues, skeletal muscle and the liver (7, 11, 12). This expression pattern suggests that adipose tissue may be the primary target for the insulin-sensitizing effect of PPARγ agonists such that changes in adipose tissue activity lead to improved insulin sensitivity in other tissues (13). Indeed, TZDs markedly change gene expression in adipose tissue, yielding smaller adipocytes whose altered secretion of adipokines may improve insulin resistance in other tissues (13, 14). Study of the necessity of adipose tissue for the antidiabetic actions of TZDs using two lipodystrophic mouse models has yielded conflicting results (15, 16). There is also evidence that TZDs directly improve insulin-stimulated glucose disposal in cultured muscle cells (17, 18).
To elucidate the physiologic role of muscle PPARγ and its significance in the actions of TZDs, we generated mice with muscle-specific deletion of PPARγ. We find that these mice develop excess adiposity as well as whole-body insulin resistance. Tissue-specific insulin sensitivity in these mice is normal in skeletal muscle but impaired in the liver and perhaps adipose tissue, as well. Interestingly, the muscle-specific PPARγ KO mice respond normally to the TZD rosiglitazone (RSG). These data suggest that muscle PPARγ is critical for maintenance of whole-body responsiveness to insulin, but is not required for the antidiabetic actions of TZDs.
Methods
Animals.
PPARγ-loxP mice on a 129/sv, C57BL/6, FVB mixed background (19) were crossed with mice harboring Cre DNA recombinase under the control of the muscle creatinine kinase promoter (MCK-Cre mice) on a 129/sv, C57BL/6, DBA mixed background (20). Animals were housed in pathogen-free facilities, maintained on a 12-hour light/dark cycle, and fed standard rodent chow (Purina Mills Inc., St. Louis, Missouri, USA) containing 8% of fat by weight unless specified. All animal protocols were approved by the Animal Care Use Committees of the Joslin Diabetes Center or Brandeis University, in accordance with NIH guidelines. In some experiments, mice were fed a high-fat diet (Harlan Teklad Laboratory, Madison, Wisconsin, USA) that contained 29.3% fat, 25.2% protein, and 28.8% carbohydrate by weight. In addition, specific groups of high-fat diet–fed mice were simultaneously given RSG (GlaxoSmithKline, Philadelphia, Pennsylvania, USA) at 3 mg/kg body weight per day as food admixture.
Mice with five genotypes were generated in this study as follows. Control mice (two intact PPARγ alleles) included WT (no PPARγ-loxP or MCK-Cre alleles); Lox (two PPARγ-loxP, no MCK-Cre); and Cre mice (no PPARγ-loxP, MCK-Cre positive). Two other groups were expected to have partial or complete PPARγ deficiency in skeletal muscle; heterozygous (Het) and KO (MuPPARγKO) mice carried one or two PPARγ-loxP alleles, respectively, and they all were MCK-Cre positive. The studies herein were performed on mice of the F2 and subsequent generations.
Genotyping and assessment of recombination of PPARγ DNA.
Genotyping for the PPARγ-loxP allele and MCK-Cre transgene was performed by PCR on DNA isolated from tail clips using described primers and conditions (20, 21), respectively. Cre-mediated recombination and deletion of exon 2 of the PPARγ gene was detected by PCR using described primers (21). Recombination and deletion of exon 2 was quantitatively assessed by Southern blot analysis of BamHI-digested high-molecular-weight genomic DNA prepared from whole muscle. Probe 1 (Figure 1a) was hybridized to depurinated DNA after alkaline transfer (22) to nylon membranes.
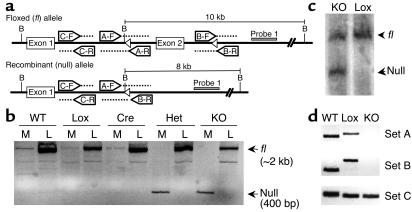
Figure 1.
Recombination of PPARγ-loxP allele in the muscle of MuPPARγKO mice. (a) Schematic of recombinant mouse PPARγ alleles indicating PCR primers sets, BamHI restriction sites (labeled as B), loxP sites (open triangles), and Probe 1 for Southern blot analysis. (b) Detection of intact (fl) versus recombined (null) allele by PCR on genomic DNA from muscle (M) or liver (L). The fl and null alleles appear at approximately 2 kb and 400 bp, respectively. (c) Southern blot analysis of BamHI-digested genomic DNA isolated from muscle samples from Lox and MuPPARγKO (KO) mice. Hybridization was performed with Probe 1, identifying fragments of 10 and 8 kb for fl and null alleles, respectively. (d) PCR detection of unrecombined alleles (primer sets A and B) or any WT, fl, or null allele (primer set C) in genomic DNA isolated from enriched myocytes of WT, Lox, and KO mice. Primer sets A and B yield larger products in the fl compared with WT allele due to loxP insertion.
To elucidate the cellular component responsible for PPARγ-loxP recombination, an established approach (23) was used to obtain myocyte-enriched fractions from skeletal muscle digested with 2 mg/ml collagenase (C-9891; Sigma-Aldrich, St. Louis, Missouri, USA), 2 mg/ml dispase (17105-041; Invitrogen Corp., Carlsbad, California, USA), and 1 mg/ml hyaluronidase (H-3506; Sigma-Aldrich). Semiquantitative PCR was performed on genomic DNA isolated from enriched striated cells pelleted at 800 g for 5 minutes using the following primers: set A (A-F, 5′ primer: 5′-CCTTTCTTCTTCTCCCTCCAA-3′; A-R, 3′ primer: 5′-CAAGGAAACAGCACCACAGA-3′); set B (B-F, 5′ primer: 5′-CAAAGGCATGGGGTCACTT-3′; B-R, 3′ primer: 5′-GGACAGCATATCCCTAACTTTCT-3′); and set C (C-F, 5′ primer: 5′-CTGTGGGAGGCCAAGATAAG-3′; C-R, 3′ primer: 5′-CCAAAGATGGAAACAAGAAAGG-3′) (Figure 1a).
Blood glucose, insulin, triglyceride, and free fatty acid measurement.
Tail blood of random-fed mice, which had free access to food, was collected at 7 to 9 hours into the light cycle. Fasting blood samples were obtained 2 to 3 hours into the light cycle after food restriction for 14 hours. Blood and plasma glucose were measured using a Glucometer Elite (Bayer Corp., Elkhart, Indiana, USA). Plasma insulin was measured with an ELISA kit (Crystal Chem, Downers Grove, Illinois, USA). In some studies, blood samples from random-fed mice were collected by retro-orbital bleeding under light anesthesia. Plasma triglyceride and FFA were assayed using commercial kits (Sigma-Aldrich and Wako Chemicals USA Inc., Richmond, Virginia, USA, respectively).
Glucose tolerance test and insulin tolerance test.
For glucose tolerance tests (GTTs), mice were fasted overnight for 14 hours followed by intraperitoneal glucose injection (2 g/kg body weight). Blood glucose was measured by tail bleeding at 0, 15, 30, 60, and 120 minutes after the injection. In some experiments, 15 μl of tail blood was collected for insulin assay at 0, 15, and 30 minutes. For insulin tolerance tests (ITTs), mice were injected with human regular insulin (Humulin; Eli Lilly and Co., Indianapolis, Indiana, USA) at 1–1.5 U/kg body weight intraperitoneally, and blood glucose was measured at 0, 15, 30, and 60 minutes.
Hyperinsulinemic-euglycemic clamp with glucose flux analysis.
A catheter (Micro-Renathane 0.025-inch outer diameter; Braintree Scientific Inc., Braintree, Massachusetts, USA) was secured in the jugular vein of mice anesthetized by intraperitoneal injection with 0.015 ml/kg of a 2.4% solution of 1:1 mixture of 2,2,2-tribromoethanol and t-amyl alcohol. Hyperinsulinemic-euglycemic clamp studies were conducted on postoperative days 3–6 in awake, tail-restrained mice after a 5-hour fast. Whole-body glucose flux was traced using a continuous infusion of [3-3H] glucose (NEN Life Science Products Inc., Boston, Massachusetts, USA) at 0.05 μCi/min after an initial 5-μCi bolus. After basal sampling, a primed (40 mU/kg) continuous infusion (4 mU/kg/min) of human regular insulin (Humulin; Eli Lilly and Co.) was administered in 0.1% BSA. Tail-tip blood samples (2 μl) were collected at 10-minute intervals. Aqueous 50% glucose was infused at variable rates as needed to maintain blood glucose at approximately 120 mg/dl.
Tissue-specific insulin-stimulated glucose transport was estimated from a bolus (10 μCi) of 2-deoxy-D-[1-14C] glucose (2-[14C]DG; NEN) administered 45 minutes before the end of the clamp. Blood samples (20 μl) were obtained at 10-minute intervals for the determination of plasma [3H] glucose and 2-[14C]DG concentrations. Additional blood samples (10 μl) were collected both during the basal period and at the end of the clamp for plasma insulin assay. At the end of the clamp, animals were euthanized, with tissues rapidly dissected and frozen in liquid N2. Plasma [3-3H] glucose and 2-[14C]DG concentrations were determined by a dual-channel scintillation counter (Beckman Coulter Inc., Fullerton, California, USA) after deproteinization and drying. Tissue 2-[14C]DG and 2-[14C]DG-6-phosphate (2-[14C]DG-6P) were separated by ion-exchange chromatography.
The rate of whole-body glucose turnover was determined at steady state from the ratio of the [3H] glucose infusion rate to the measured specific activity of plasma glucose (24). Hepatic glucose production (HGP) was determined by subtraction of the glucose infusion rate from the whole-body glucose turnover (25). Tissue-specific glucose transport was calculated from tissue 2-[14C]DG-6P content normalized against the area under the plasma 2-[14C]DG decay curve (26).
Transport of 2-deoxy-glucose in isolated skeletal muscle.
Control (Lox) and MuPPARγKO mice were fed a high-fat diet for 7 weeks initiated at 4 months of age. Following a 10- to 12-hour fast, mice were sacrificed, and soleus and extensor digitorum longus (EDL) muscles were rapidly and carefully dissected, tied with suture (silk 4-0), and mounted on an incubation apparatus with resting tension of 0.25 g (27). Muscles were preincubated in Krebs-Ringer bicarbonate buffer (KRB: 117 mM NaCl, 4.7 mM KCl, 2.5 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, 24.6 mM NaHCO3, pH 7.4) containing 2 mM pyruvate at 37°C for 40 minutes. To measure 2-deoxy-glucose (2-DG) uptake, muscles were transferred to 2 ml KRB containing 1 mM 2-deoxyglucose and 2-deoxy-D-[1, 2-3H]-glucose (2-[3H]DG, 1.5 μCi/ml), 1 mM D-mannitol, and D-[14C]-mannitol (0.45 μCi/ml) at 30°C for 10 minutes. Basal uptake of 2-[3H]DG was measured in the absence of insulin, while uptake in the contralateral muscle was measured in the presence of a submaximal insulin concentration (6 nM human insulin). Insulin-stimulated muscle 2-DG uptake was determined as described previously (20).
Carcass analysis, fat pad weight, and tissue triglyceride measurement.
Analysis for body triglyceride content was performed by carcass saponification in ethanolic KOH as described (28), except glycerol content was measured (Kit 227B; Sigma-Aldrich) after neutralization with MgCl2. Tibialis anterior muscle and hepatic triglyceride content were determined by ethanolic KOH saponification followed by assay for glycerol.
RNA analysis.
RNA was extracted from mixed hindlimb skeletal muscle using TRIzol (Life Technology Inc., Rockville, Maryland, USA). For Northern blot analysis, 10 μg of RNA was transferred from denaturing agarose gel onto Bright Star-Plus nylon membranes (Ambion Inc., Austin, Texas, USA) using QuickHyb (Stratagene, Cedar Creek, Texas, USA). Hybridized membranes were exposed to a PhosphorImage Screen (Molecular Dynamics, Sunnyvale, California, USA), analyzed using ImageQuant software (Molecular Dynamics), and normalized to the expression of 36B4. The cDNAs were synthesized using Superscript II reverse transcriptase (Life Technologies Inc.) and oligo(dT) priming. Probes for specific cDNAs were prepared by PCR using Taq polymerase (Life Technologies Inc.) and mouse skeletal muscle cDNA with the following primers: lipoprotein lipase (LPL: 5′ primer, 5′-GACACAGCTGAGGACCATTGTCAT-3′; 3′ primer, 5′-GCAGCATGGGCTCCAAGGCTGTA-3′), heart/muscle fatty acid–binding protein (FABP3; 5′ primer, 5′-GGCGGACGCCTTTGTCGGTACCT-3′; 3′ primer, 5′-GGATGAGTTTCCCGTCAACTAGCT-3′), and muscle mitochondrial carnitine palmitoyltransferase-1 (mCPT1b; 5′ primer, 5′-GGCAACAGTTGGTTCCAACTACT-3′; 3′ primer, 5′-CAGGAAGCTTAGGCATGTACGTT-3′). The probe for 36B4 was generated using the primers described (29). All PCR products were confirmed to be free of mutation by sequencing.
For DNA microarrays, samples were prepared following the Affymetrix GeneChip Expression Analysis Manual (Affymetrix, Santa Clara, California, USA) with minor modifications. Total RNA was cleaned using an RNeasy Mini Kit (QIAGEN Inc., Valencia, California, USA). From 25–35 μg of total RNA, double-stranded cDNA was created using the SuperScript Choice system (Life Technologies Inc.). First-strand cDNA synthesis was primed with T7-(dT24) oligonucleotide. The cRNA was synthesized using T7 MegaScript in vitro Transcription Kit (Ambion Inc.), labeled with biotin using Bio-11-CTP and Bio-16-UTP (Enzo Diagnostics Inc., Farmingdale, New York, USA), and cleaned using RNeasy Mini Kit. The cRNA was treated in fragmentation buffer (40 mM Tris-acetate, pH 8.1, 100 mM KOAc, 30 mM MgAc) for 35 minutes at 94°C. Fragmented cRNA (27 μg) was hybridized on murine U74Av2 arrays for 24 hours in a 45°C hybridization oven, stained with streptavidin-phycoerythrin (Molecular Probes Inc., Eugene, Oregon, USA) in Affymetrix fluidics stations, scanned fluorometrically (Hewlett Packard Corp., Palo Alto, California, USA), and analyzed using GENECHIP MAS V.4.0.
Statistics.
Results were expressed as mean plus or minus SEM. Statistical analysis was performed using a two-tailed unpaired t test for dual samples and ANOVA for groups. For expression array analysis, a gene was considered significantly altered between groups if each of the following three conditions were met: (a) the expression intensity in at least one of the groups was greater than 3 × noise + background; (b) the difference in expression intensity between groups was greater than 3 × noise; and (c) the two-tailed unpaired t test between groups yielded P values less than 0.05.
Results
Creation of MuPPARγKO mice.
To study the functional role of PPARγ in skeletal muscle, we conditionally disrupted the PPARγ gene in skeletal muscle using Cre/loxP–mediated DNA recombination (30) by crossing PPARγ-loxP mice with MCK-Cre mice. In the PPARγ-loxP mice, loxP sites were introduced both upstream and downstream of exon 2 of the PPARγ gene (19). In the presence of Cre recombinase, exon 2 of the PPARγ-loxP allele is deleted, resulting in a message containing a frameshift and premature stop codon after exon 1. In published tissue-specific KOs using this allele, the resultant PPARγ transcript is indeed truncated with reduced message half-life, and no PPARγ protein can be detected (19, 31). The muscle-specific expression and functional activity of Cre DNA recombinase under control of the MCK promoter have been described (20, 32).
Recombination of the PPARγ-loxP allele in tissues was detected by PCR, amplifying an approximately 2-kb product from the intact PPARγ-loxP allele (fl) and an approximately 400-bp product from the recombined PPARγ-loxP allele (null). As expected, recombination of the PPARγ gene was present only in the skeletal muscle of Het and KO, but not in WT, Cre, and Lox mice (Figure 1b). Recombination of the PPARγ gene was specific for skeletal muscle and was not observed in the liver. Expression of PPARγ protein in adipose tissue was normal in MuPPARγKO mice as measured by Western blot analysis (data not shown).
Measurement of the specific loss of PPARγ from skeletal muscle was complicated by contamination of skeletal muscle preparations with some adipose tissue (33). Because levels of PPARγ are many fold higher in adipocytes than in skeletal muscle, even low-level contamination is a significant confounder to accurate quantitation of muscle PPARγ transcript or protein levels. Knowing that deletion of exon 2 from the PPARγ-loxP allele prevents PPARγ expression, we quantitatively approached this problem by measuring the loss of exon 2 in genomic DNA prepared from skeletal muscle, thus avoiding the above relative-expression quandary. Deletion of exon 2 was measured by Southern blot analysis of BamHI-digested genomic DNA prepared from whole muscle tissue. Hybridization of probe 1 at approximately 8 kb, corresponding to the recombined PPARγ-loxP allele (null), was detected in muscle of MuPPARγKO but not Lox (Figure 1c) or WT mice (not shown). In muscle from Lox mice, only the unrecombined PPARγ-loxP (fl) allele, at approximately 10 kb, was detected. As expected, the unrecombined allele was also detected in DNA prepared from MuPPARγKO muscle, such that the recombined allele represented 50% ± 0.3% (n = 3) of muscle DNA. This frequency is near that expected for complete myocyte-specific excision in muscle, because myocytes make up 50% or less of this tissue (34–36) with the remainder made up by myoblasts, endothelium, intramural adipocytes, etc.
To confirm that myocytes in skeletal muscle are specifically responsible for the observed loss of exon 2, we prepared myocyte-enriched cellular fractions from Lox and MuPPARγKO mice. PCR was performed on genomic DNA isolated from the myocyte-enriched fraction to detect portions of the PPARγ-loxP allele that are preserved (primer set C) or disrupted (primer sets A and B) by Cre-mediated excision (see primer schematic in Figure 1a). As expected, we were able to readily detect the nondisrupted part of the allele in WT, Lox, and MuPPARγKO myocytes using primer set C (Figure 1d). The products from primer sets A and B were readily detected in WT and Lox myocyte DNA, with the expected change in size, but were not detected in MuPPARγKO myocyte DNA, even after 45 cycles of amplification (Figure 1d). In addition, we detected the null allele using primers spanning the deletion site only in myocytes from MuPPARγKO mice (data not shown). Taken together, these data indicate extensive and specific disruption of PPARγ in myocytes of MuPPARγKO mice.
Effect of muscle deletion of PPARγ on growth, body composition, and lipid distribution.
The growth of MuPPARγKO, Lox, and control mice fed normal chow was followed for 12 months, with no observed differences (Figure 2a). We found, however, that both Het and MuPPARγKO had significantly enlarged epididymal fat pads compared with controls (Figure 2b; P < 0.05). This was accompanied by a small but nonsignificant increase in serum leptin levels in Het and MuPPARγKO mice compared with control mice (control: 4.7 ± 0.9 ng/ml; Het: 5.6 ± 1.0 ng/ml; KO: 6.2 ± 1.2 ng/ml).
Figure 2.
Excess adiposity in MuPPARγKO mice exacerbated by high-fat diet. (a) Body mass of male mice on normal diet: WT (open squares), Cre (open diamonds), Lox (open triangles), Het (shaded circles), and MuPPARγKO (filled triangle). (b) Epididymal fat pad mass of 4-month-old mice fed normal diet. (c) Body mass of male mice following switch from normal to high-fat diet at 4 months of age: Lox (open triangles) and MuPPARγKO (filled triangles). (d) Whole-body triglyceride content of mice following 7 weeks of high-fat diet initiated at 4 months of age. (e) Daily intake of food per male mouse measured during weeks 3–7 following switch from normal to high-fat diet at 4 months of age. *P < 0.05 versus control; #P < 0.01 versus control.
When fed a high-fat diet, MuPPARγKO mice gained significantly more weight than control (Lox) mice (Figure 2c). Whole-body triglyceride content of MuPPARγKO mice fed 7 weeks of high-fat diet was 51% higher (P = 0.05) than that of control mice (Figure 2d), accounting for the excess body mass in the MuPPARγKO mice. The excess weight gain in MuPPARγKO mice compared with Lox controls occurred despite a significant and persistent reduction in food intake for mice on the high-fat diet (Figure 2e).
Given the excess lipid deposition in adipose tissue in MuPPARγKO mice, we examined other lipid parameters. We found similar serum FFA levels in MuPPARγKO, Het, and control mice on a regular diet (1.37 ± 0.08, 1.25 ± 0.05, and 1.21 ± 0.05 mEq/l, respectively) or a high-fat diet at 6 months of age. Circulating triglyceride levels were not different between MuPPARγKO, Het, and control mice (113 ± 14, 121 ± 13, and 121 ± 13 mg/dl, respectively). Triglyceride content of skeletal muscle was also not different between MuPPARγKO, Het, and control mice (8.4 ± 1.0, 8.2 ± 1.2, and 8.0 ± 0.9 mg/g, respectively). In addition, there was normal liver triglyceride content in MuPPARγKO compared with Lox mice (11.5 ± 1.2 versus 13.6 ± 2.8 mg/g).
Glucose homeostasis in MuPPARγKO mice.
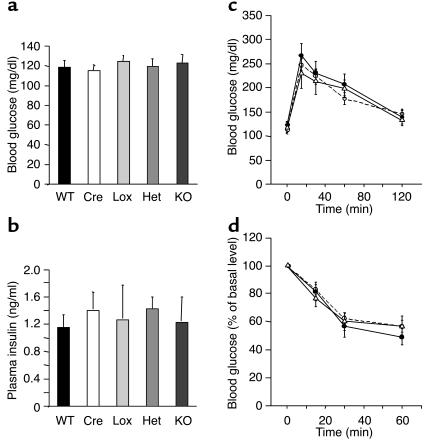
To assess the effect of the muscle PPARγ KO on whole-body glucose homeostasis, we measured glucose and insulin levels, GTT, and ITT in mice fed normal chow. We found no differences in random glucose and insulin levels in either Het or MuPPARγKO mice as compared with WT, Cre, and Lox control mice (Figure 3, a and b). In addition, MuPPARγKO and Het mice displayed normal response to GTT and ITT, as compared with control mice (Figure 3, c and d). We detected no differences in response patterns of male and female mice (data not shown).
Figure 3.
MuPPARγKO mice have normal glucose and insulin levels, as well as normal response to glucose or insulin challenge. (a) Random-fed blood glucose of 4-month-old male mice. (b) Random-fed insulin levels of 4-month-old male mice. (c) GTT (2 g glucose/kg, intraperitoneal) of 4-month-old male mice after overnight fast. Control mice (includes WT, Cre, and Lox: open circles); Het (filled circles); KO (open triangles). (d) ITT (1 U insulin/kg, intraperitoneally) of 4-month-old male mice. Control mice (includes WT, Cre, and Lox: open circles); Het (filled circles); KO (open triangles).
Insulin resistance in MuPPARγKO mice due to impaired hepatic insulin action.
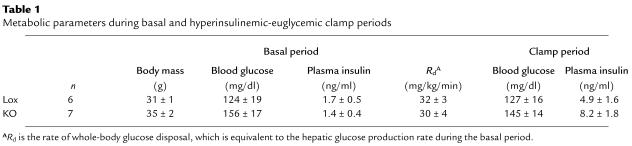
The effect of muscle PPARγ KO on insulin sensitivity was directly assessed by 2-hour hyperinsulinemic-euglycemic clamp in conscious MuPPARγKO 4-month-old male mice, using sibling Lox mice as controls. Baseline blood glucose and plasma insulin levels were not statistically different between the two genotypes during the basal period of the clamp (Table 1), though there was a weak tendency toward higher basal glucoses in the MuPPARγKO mice. The increase in plasma insulin levels during the clamp was not significantly different between the two genotypes (Table 1; P = 0.25), and euglycemia was maintained in both groups. Both genotypes had nearly identical glucose disposal rates (Rd) during the basal period (Table 1). MuPPARγKO mice, however, required a 36% lower glucose infusion rate (28 ± 4 versus 44 ± 5 mg/kg/min in controls; P < 0.05, n = 6–7) (Figure 4a) to maintain euglycemia during the hyperinsulinemic portion of the clamp compared with Lox controls, demonstrating whole-body insulin resistance in the MuPPARγKO mice.
Table 1.
Metabolic parameters during basal and hyperinsulinemic-euglycemic clamp periods
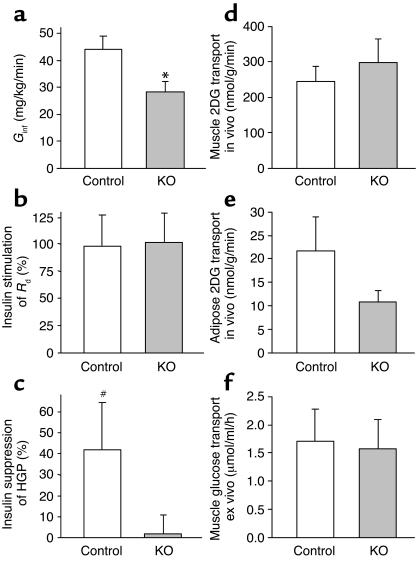
Figure 4.
Whole-body insulin resistance due to impaired insulin action in liver not skeletal muscle. (a) Glucose infusion rate (Ginf) required to maintain euglycemia during hyperinsulinemic clamp in 4-month-old male MuPPARγKO and control (Lox) mice. (b) Insulin stimulated increase in the Rd during hyperinsulinemic compared with basal portion of clamp. (c) Insulin stimulated the percentage suppression of HGP during hyperinsulinemic compared with basal portion of the clamp. Insulin stimulated 2-[14C]DG (2DG) uptake in vivo during hyperinsulinemic-euglycemic clamp in 4-month-old male MuPPARγKO and control (Lox) mice, in mixed hindquarter muscle (d), or in adipose tissue (e). (f) Insulin stimulated 2-[3H]DG uptake above basal in soleus muscle isolated ex vivo from MuPPARγKO and control (Lox) male mice fed a high-fat diet initiated at 4 months of age for 7 weeks. *P < 0.05 versus control; #P < 0.05 versus null hypothesis of no suppression.
The defect in whole-body insulin sensitivity was not due to changes in glucose disposal, because the MuPPARγKO mice had an identical insulin-stimulated increase in Rd compared with controls (Figure 4b). In contrast, there was a marked defect in insulin action in the livers of MuPPARγKO mice such that there was a complete failure of insulin to suppress HGP in these mice, while in control mice there was a significant suppression of HGP by insulin (Figure 4c; P = 0.03). Thus, somewhat surprisingly, hepatic insulin resistance appears to account for the whole-body insulin resistance observed in MuPPARγKO mice.
Normal insulin-stimulated glucose-disposal in skeletal muscle of MuPPARγKO mice.
Skeletal muscle and adipose tissue both contribute to insulin-stimulated Rd, with skeletal muscle accounting for the majority of disposal (3). The normal insulin-stimulated increase in Rd in MuPPARγKO mice suggested that insulin-stimulated glucose uptake in skeletal muscle of these mice was normal. To test this directly and deconvolute tissue-specific determinants of glucose uptake, we measured the uptake of a bolus of 2-[14C]DG given during the hyperinsulinemic portion of the clamp. As expected from the above Rd data, transport of 2-[14C]DG was not impaired in the skeletal muscle of MuPPARγKO mice (Figure 4d). There was a trend, however, toward reduced uptake of 2-[14C]DG in the epididymal fat of the MuPPARγKO mice (Figure 4e).
As independent evaluation of insulin action in skeletal muscle, we measured 2-[3H]DG uptake in isolated individual muscle groups isolated from MuPPARγKO and control (Lox) mice. To promote insulin resistance, both groups were fed a high-fat diet for 7 weeks preceding the measurement. Consistent with the tissue-specific clamp results, the insulin-stimulated 2-DG uptake above basal in soleus muscle (Figure 4f) was unaffected in MuPPARγKO mice. The insulin-stimulated uptake above basal of 2-DG in EDL was also not significantly changed in MuPPARγKO animals (0.8 ± 0.5 versus 0.4 ± 0.1 μmol/ml/h in Lox controls).
Glucose homeostasis response to high-fat diet and RSG in MuPPARγKO mice.
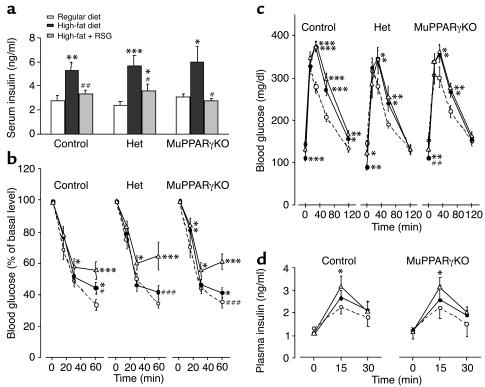
Control, Het, and MuPPARγKO mice were fed either a regular or high-fat diet for 2–4 weeks. In control mice, ingestion of the high-fat diet did not cause hyperglycemia (data not shown), but dramatically increased plasma insulin levels (Figure 5a). The mice on high-fat diet exhibited an impaired response to intraperitoneal insulin (Figure 5b) as well as impaired glucose tolerance (Figure 5c). Het and MuPPARγKO mice showed a response that was indistinguishable from that of controls with respect to induction of hyperinsulinemia, impaired intraperitoneal insulin response, and glucose intolerance on the high-fat diet challenge (Figure 5, a–c).
Figure 5.
RSG improves glucose homeostasis in MuPPARγKO mice fed a high-fat diet. (a) Random-fed insulin levels after 2 weeks of regular diet, high-fat diet, or high-fat diet plus RSG, in control, Het, and MuPPARγKO mice. (b) ITT (1.5 U/kg, intraperitoneally) of mice treated with regular diet, high-fat diet, or high-fat diet plus RSG for 4 weeks. (c) GTT (2 g/kg, intraperitoneally) after overnight fast. (d) Serum insulin concentrations during GTT. Groups: regular diet (open circles), high-fat diet (open triangles), and high-fat diet plus RSG (filled circles). *P < 0.05 versus regular diet; **P < 0.01 versus regular diet; ***P < 0.001 versus regular diet; #P < 0.05 versus high-fat diet; ##P < 0.01 versus high-fat diet; ###P < 0.001 versus high-fat diet.
To study the role of muscle PPARγ in mediating the effects of TZDs, the high fat–fed mice were simultaneously treated with RSG for 2–4 weeks. In control mice, RSG treatment was largely able to prevent the high fat–induced abnormalities of plasma insulin levels (Figure 5a) and of the response to intraperitoneal insulin (Figure 5b). Although there was minimal effect of RSG on GTT (Figure 5c), the RSG-treated mice tended to have lower induced insulin levels as compared with mice on high-fat diet alone during GTT (Figure 5d), indicating that the hypoinsulinemic effect of RSG was not affected by deficiency of PPARγ in skeletal muscle.
Expression of genes involved in lipid metabolism.
Given the abnormality in lipid distribution resulting from muscle-specific disruption of PPARγ, we explored the expression in skeletal muscle of a wide set of lipid metabolism genes using Affymetrix expression microarrays profiling 12,488 genes. We identified and screened approximately 100 lipid metabolism genes for significant alteration in MuPPARγKO compared with Lox control mice. Two of these were significantly altered (Figure 6a); fatty acid transport protein-1 (FATP1) and long-chain fatty acyl-coA synthetase 2 (LC-FACS2) were each significantly upregulated by 3.21-fold ± 0.39-fold and 1.74-fold ± 0.20-fold, respectively. Of note, the expression of PPARα and PPARδ were not significantly altered in MuPPARγKO mice, as measured by microarray analysis.
Figure 6.
Expression of lipid metabolism genes in skeletal muscle of MuPPARγKO mice. (a) Expression of lipid metabolism genes calculated from the average of three microarrays for six mice for each genotype. The data are presented as the ratio of intensity between MuPPARγKO and Lox mice. Standard errors were estimated using the zero method, for the purpose of display only. (b) Northern blot of RNA extracted from hindlimb muscles. *Statistically altered genes as determined by screening criteria detailed in Methods. #P < 0.05 versus control, based on Northern blot analysis.
Previous work has found a correlation between muscle PPARγ levels and the expression of three lipid metabolism genes: mCPT1b, FABP3, and LPL (37). Microarray analysis showed no change in expression of LPL (fold = 1.08) and a trend toward upregulation of FABP3 in MuPPARγKO (fold = 1.39) (Figure 6a). Mouse mCPT1b is not represented on the U74Av2 microarray. Northern blot analysis of muscle RNA was used to independently explore the role of muscle PPARγ in the expression of these three lipid metabolism genes (Figure 6b). There was a significant increase in FABP3 message in the skeletal muscle of MuPPARγKO mice nearly identical to that found by microarray profiling, with a fold change of 1.47 reaching statistical significance (P < 0.05). Likewise, there was close agreement between Northern and microarray analysis for LPL, with an insignificant fold change of 1.12. Northern blot analysis also showed no statistical regulation of mCPT1b (fold = 1.13).
Discussion
The contribution of PPARγ activity in specific tissues to whole-body insulin sensitivity has been difficult to dissect. Although PPARγ levels are highest in white adipose tissue, multiple insulin-sensitive tissues express PPARγ and thus in theory might contribute to the effects of TZDs on whole-body insulin sensitivity. Furthermore, the endogenous role of PPARγ in tissues has been difficult to determine since global genetic KO of PPARγ in the mouse results in embryonic lethality by embryonic day 10 (38), and mice with heterozygous deficiency of PPARγ display improved insulin sensitivity (39, 40). In humans, specific PPARγ mutations have been shown to lead to obesity (41) or marked insulin resistance (42), and a common polymorphism (Pro12Ala) is associated with improved insulin sensitivity (43, 44). To explore the role of muscle PPARγ in insulin sensitivity and glucose homeostasis, we generated muscle-specific PPARγ–deficient mice using a Cre/loxP strategy.
We find that mice with muscle-specific disruption of PPARγ have normal glucose homeostasis and insulin levels, including normal responses to glucose or pharmacologic insulin challenge. The MuPPARγKO mice, however, have significant whole-body insulin resistance when measured directly using the hyperinsulinemic-euglycemic clamp. This mild insulin-resistance phenotype is similar to that of the muscle-specific insulin receptor KO (MIRKO) mouse, which has normal glucose and insulin levels and normal responses to glucose or insulin challenge (20), yet has insulin resistance when measured by euglycemic-hyperinsulinemic clamp (26). In MIRKO mice, this is associated with severe impairment of insulin-stimulated glucose uptake in muscle and compensatory increased glucose uptake in fat due to undefined tissue crosstalk (26). In contrast, in MuPPARγKO mice the whole-body insulin resistance is localized to nonmuscle tissues, including the liver and, possibly, fat.
Related results have been observed in mice with selective KO of glucose transporter-4 in muscle (M-G4KO). These mice have severe loss of basal, insulin-stimulated, and contraction-stimulated glucose uptake in skeletal muscle (32) and have whole-body insulin resistance as measured by euglycemic-hyperinsulinemic clamp (45). The glucose homeostasis defect in M-G4KO mice is more severe, and they develop secondary insulin resistance in liver and adipose tissue mediated by glucose toxicity (45). Since MuPPARγKO mice have normal glucose levels and normal insulin-stimulated glucose uptake in muscle, it is likely that a mechanism other than glucose toxicity is responsible for the insulin resistance that develops in the liver and, possibly, fat of these mice.
The hepatic insulin resistance in MuPPARγKO mice might be secondary to altered adipokine release associated with increased adiposity. Consistent with this possibility, preliminary studies suggest altered adipokine expression in adipose tissue as determined by quantitative RT-PCR. The expression of ACRP30 was reduced 55% in adipose tissue from MuPPARγKO mice compared with controls (P = 0.09), while the expression of TNF-α was increased 63% (P = NS) in adipose tissue from MuPPARγKO mice (unpublished data). Both of these trends could be expected to induce hepatic insulin resistance (46, 47), though proof of causality in our model will require additional experimentation.
The disruption of muscle PPARγ in mice leads to excess adiposity. This phenotype is similar to that of MIRKO mice, which also have normal body weight on normal chow but develop increased fat depot mass (20). In MIRKO mice, however, this is accompanied by increased insulin-stimulated glucose uptake in adipose tissue. Since MuPPARγKO mice have a tendency for diminished responsiveness to insulin in adipose tissue, it is unlikely that MIRKO and MuPPARγKO mice develop enlarged fat pads via the same mechanisms.
The excess weight gain and adiposity observed in MuPPARγKO mice on a high-fat diet occurred despite a reduction in caloric intake. This suggests that the adiposity in MuPPARγKO mice is not due to a defect in appetite regulation such as would occur in leptin resistance. In fact, the decrement in food intake associated with increased adiposity, coupled with the observed trend toward higher serum leptin levels, is consistent with an intact leptin response in MuPPARγKO mice. The feeding data instead suggest that the weight gain/adiposity of MuPPARγKO mice may be due to higher energy efficiency, that is, reduced fuel oxidation and increased fuel storage. Given that skeletal muscle is a major site of fuel oxidation (48), it is possible that loss of muscle PPARγ could produce such an effect. A preliminary indirect calorimetry study has revealed no differences in oxygen consumption, CO2 production, and physical activity levels between MuPPARγKO and Lox control mice on a high-fat diet (unpublished data). Likewise, we found no significant differences in uncoupling protein-1, -2, or -3 expression between MuPPARγKO and Lox muscle, as represented on the U74Av2 microarray (unpublished data).
An alternative explanation for excess fat mass in MuPPARγKO mice would be an impaired ability of skeletal muscle to use lipid fuel substrate leading to a shunting of lipid to adipose tissue. Indeed, it is postulated that PPARγ mediates upregulation of lipid use in skeletal muscle (49–51), in a manner similar to the essential role of isoform PPARα in the upregulation of lipid oxidation in cardiac muscle (52, 53). Though isoform PPARα is expressed in skeletal muscle at levels higher than PPARγ (ref. 54; our unpublished data), there is little to no impairment in skeletal muscle lipid utilization gene regulation upon loss of PPARα (55). This suggests that other factors, perhaps PPARγ, play an important role. Consistent with this possibility, there are differences in lipid metabolism gene expression in MuPPARγKO muscle.
Defects in use of fatty acids by skeletal muscle have also been postulated to contribute to the development of type 2 diabetes (56, 57), such that excess delivery of lipid to liver and adipose tissues leads to insulin resistance in these tissues (58). This represents an alternative mechanism that could account for the diminished insulin sensitivity in liver and adipose tissue in MuPPARγKO mice, though we did not detect differences in lipid content of serum or tissues of MuPPARγKO mice other than increased adipose mass.
TZDs improve whole-body glucose homeostasis by increasing insulin-stimulated glucose uptake in skeletal muscle (5–7). This action on muscle may be direct (16–18) or indirect through the effects of TZDs on fat (13–15). It is thus informative that although MuPPARγKO mice develop insulin resistance at baseline, they nonetheless have a normal response to RSG with a reduction in hyperinsulinemia and improved whole-body glucose homeostasis. Taken together, these improvements are very suggestive of increased insulin sensitivity, though in the absence of clamp data from TZD-treated MuPPARγKO mice it remains possible that subtle deficiencies in the TZD response of MuPPARγKO mice exist. The normal response in multiple parameters of MuPPARγKO mice to TZDs, however, supports the hypothesis that PPARγ agonists improve insulin sensitivity by direct actions on tissues other than skeletal muscle, presumably adipose tissue. This hypothesis has been suggested in a previous study where TZDs failed to improve glucose homeostasis in mice with severe lipoatrophy (15). Adipose tissue secretes a number of factors, including leptin, TNF-α, FFAs, resistin, IL-6, and ACRP30, which modulate insulin sensitivity (59). TZDs can induce beneficial changes in the adipose production of most of these, including lower FFAs (60), TNF-α (61), and resistin (62) levels, as well as increased ACRP30 levels (63). These salutary changes are accompanied by a reduction of adipocyte size, which is also expected to improve overall body-glucose homeostasis (64). Any or a combination of these factors could represent a mechanism for the insulin-sensitizing effects of TZDs on skeletal muscle by adipose tissue.
Our data, as well as other recent studies, suggest that PPARγ in skeletal muscle may directly modulate expression of some genes of lipid metabolism. In normal human skeletal muscle, Lapsys et al. (37) found a strong positive correlation between the expression level of PPARγ and the mRNA levels of LPL, FABP3, and mCPT1b. In contrast, we observed an increase of FABP3 in MuPPARγKO mice, but no change in LPL or mCPT1b. We did observe, however, changes in expression of LC-FACS2 and FATP1 in skeletal muscle of MuPPARγKO mice. Both of these genes are regulated in response to PPAR activity in liver (65), but in a positive manner — the opposite of the effect we observed. These findings reinforce the suggestion by Way et al. (51) that PPARγ activity in skeletal muscle may have different effects on gene expression compared with adipose tissue or liver. An in-depth analysis of gene expression in muscle of these mice before and after TZD treatment should allow definition of the genes regulated by PPARγ in skeletal muscle.
In summary, disruption of muscle PPARγ leads to increased fat mass, as well as hepatic insulin resistance. Disruption of muscle PPARγ, however, does not block the beneficial effects of TZDs on glucose homeostasis. These data indicate that the insulin-sensitizing effects of TZDs on muscle are indirect, but that PPARγ in muscle plays a role in regulation of whole-body lipid storage and hepatic insulin sensitivity through tissue crosstalk, perhaps mediated by alterations in regulation of muscle lipid metabolism genes.
Acknowledgments
We greatly appreciate the technical assistance of Elizabeth Fletcher, Scott Lannon, and Laureen Mazzola. We thank Vijay Yechoor for helpful advice. This work was supported by NIH grants DK31036 and DK45935, Joslin Diabetes and Endocrinology Research Center grant DK36836, and training grant T32-DK63702-01 (A.W. Norris).
Footnotes
A.W. Norris and L. Chen contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: thiazolidinedione (TZD); peroxisome proliferator-activated receptor γ (PPARγ); rosiglitazone (RSG); Cre recombinase under action of the muscle creatinine kinase promoter (MCK-Cre); heterozygous (Het); muscle-specific PPARγ KO (MuPPARγKO); glucose tolerance test (GTT); insulin tolerance test (ITT); 2-deoxy-d-[1-14C] glucose (2-[14C]DG); 2-[14C]DG-6-phosphate (2-[14C]DG-6P); hepatic glucose production (HGP); extensor digitorum longus (EDL); 2-deoxy-glucose (2-DG); Krebs-Ringer bicarbonate buffer (KRB); 2-deoxy-d-[1, 2-3H]-glucose (2-[3H]DG); lipoprotein lipase (LPL); fatty acid–binding protein (FABP3); muscle mitochondrial carnitine palmitoyltransferase-1 (mCPT1b); rate of glucose disposal (Rd); fatty acid transport protein-1 (FATP1); long-chain fatty acyl-coA synthetase 2 (LC-FACS2); muscle-specific insulin receptor KO (MIRKO); glucose transporter-4 KO in muscle (M-G4KO).
References
- 1.Martin BC, et al. Role of glucose and insulin resistance in development of type 2 diabetes mellitus: results of a 25-year follow-up study. Lancet. 1992;340:925–929. doi: 10.1016/0140-6736(92)92814-v. [DOI] [PubMed] [Google Scholar]
- 2.Lillioja S, et al. Insulin resistance and insulin secretory dysfunction as precursors of non-insulin-dependent diabetes mellitus: prospective studies of Pima Indians. N. Engl. J. Med. 1993;329:1988–1992. doi: 10.1056/NEJM199312303292703. [DOI] [PubMed] [Google Scholar]
- 3.DeFronzo RA, et al. The effect of insulin on the disposal of intravenous glucose: results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes. 1981;30:1000–1007. doi: 10.2337/diab.30.12.1000. [DOI] [PubMed] [Google Scholar]
- 4.Veroni MC, Proietto J, Larkins RG. Evolution of insulin resistance in New Zealand obese mice. Diabetes. 1991;40:1480–1487. doi: 10.2337/diab.40.11.1480. [DOI] [PubMed] [Google Scholar]
- 5.Petersen KF, et al. Mechanism of troglitazone action in type 2 diabetes. Diabetes. 2000;49:827–831. doi: 10.2337/diabetes.49.5.827. [DOI] [PubMed] [Google Scholar]
- 6.Kraegen EW, James DE, Jenkins AB, Chisholm DJ, Storlien LH. A potent in vivo effect of ciglitazone on muscle insulin resistance induced by high fat feeding of rats. Metabolism. 1989;38:1089–1093. doi: 10.1016/0026-0495(89)90045-0. [DOI] [PubMed] [Google Scholar]
- 7.Zierath JR, et al. Role of skeletal muscle in thiazolidinedione insulin sensitizer (PPARgamma agonist) action. Endocrinology. 1998;139:5034–5041. doi: 10.1210/endo.139.12.6364. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann JM, et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J. Biol. Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 9.Rosen ED, Spiegelman BM. PPARgamma: a nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 10.Rosen ED, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vidal-Puig AJ, et al. Peroxisome proliferator-activated receptor gene expression in human tissues. Effects of obesity, weight loss, and regulation by insulin and glucocorticoids. J. Clin. Invest. 1997;99:2416–2422. doi: 10.1172/JCI119424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruszynska YT, et al. Skeletal muscle peroxisome proliferator-activated receptor-gamma expression in obesity and non-insulin-dependent diabetes mellitus. J. Clin. Invest. 1998;101:543–548. doi: 10.1172/JCI1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spiegelman BM. PPARγ: adipogenic regulator and thiazolidinedione receptor. Diabetes. 1998;47:507–514. doi: 10.2337/diabetes.47.4.507. [DOI] [PubMed] [Google Scholar]
- 14.Hallakou S, et al. Pioglitazone induces in vivo adipocyte differentiation in the obese Zucker fa/fa rat. Diabetes. 1997;46:1393–1399. doi: 10.2337/diab.46.9.1393. [DOI] [PubMed] [Google Scholar]
- 15.Chao L, et al. Adipose tissue is required for the antidiabetic, but not for the hypolipidemic, effect of thiazolidinediones. J. Clin. Invest. 2000;106:1221–1228. doi: 10.1172/JCI11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burant CF, et al. Troglitazone action is independent of adipose tissue. J. Clin. Invest. 1997;100:2900–2908. doi: 10.1172/JCI119839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arakawa K, et al. Actions of novel antidiabetic thiazolidinedione, T-174, in animal models of non-insulin-dependent diabetes mellitus (NIDDM) and in cultured muscle cells. Br. J. Pharmacol. 1998;125:429–436. doi: 10.1038/sj.bjp.0702066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciaraldi TP, Gilmore A, Olefsky JM, Goldberg M, Heidenreich KA. In vitro studies on the action of CS-045, a new antidiabetic agent. Metabolism. 1990;39:1056–1062. doi: 10.1016/0026-0495(90)90166-a. [DOI] [PubMed] [Google Scholar]
- 19.Akiyama TE, et al. Conditional disruption of the peroxisome proliferator-activated receptor gamma gene in mice results in lowered expression of ABCA1, ABCG1, and apoE in macrophages and reduced cholesterol efflux. Mol. Cell Biol. 2002;22:2607–2619. doi: 10.1128/MCB.22.8.2607-2619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruning JC, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 21.Cui Y, et al. Loss of the peroxisome proliferation-activated receptor gamma (PPARgamma) does not affect mammary development and propensity for tumor formation but leads to reduced fertility. J. Biol. Chem. 2002;277:17830–17835. doi: 10.1074/jbc.M200186200. [DOI] [PubMed] [Google Scholar]
- 22.Wahl GM, Stern M, Stark GR. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc. Natl. Acad. Sci. U. S. A. 1979;76:3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyles GA, Archer DR. Monoamine oxidase activities in dissociated cell fractions from rat skeletal muscle. J. Pharm. Pharmacol. 1986;38:288–293. doi: 10.1111/j.2042-7158.1986.tb04568.x. [DOI] [PubMed] [Google Scholar]
- 24.Fisher SJ, et al. Determinants of glucose turnover in the pathophysiology of diabetes: an in vivo analysis in diabetic dogs. Diabetes Metab. 1996;22:111–121. [PubMed] [Google Scholar]
- 25.Fisher SJ, et al. A moderate decline in specific activity does not lead to an underestimation of hepatic glucose production during a glucose clamp. Metabolism. 1996;45:587–593. doi: 10.1016/s0026-0495(96)90028-1. [DOI] [PubMed] [Google Scholar]
- 26.Kim JK, et al. Redistribution of substrates to adipose tissue in mice with selective insulin resistance in muscle promotes obesity. J. Clin. Invest. 2000;105:1791–1797. doi: 10.1172/JCI8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi T, Hirshman MF, Kurth EJ, Winder WW, Goodyear LJ. Evidence for 5′ AMP-activated protein kinase mediation of the effect of muscle contraction on glucose transport. Diabetes. 1998;47:1369–1373. doi: 10.2337/diab.47.8.1369. [DOI] [PubMed] [Google Scholar]
- 28.Salmon DM, Flatt JP. Effect of dietary fat content on the incidence of obesity among ad libitum fed mice. Int. J. Obes. 1985;9:443–449. [PubMed] [Google Scholar]
- 29.Hill AL, Phelan SA, Loeken MR. Reduced expression of pax-3 is associated with overexpression of cdc46 in the mouse embryo. Dev. Genes Evol. 1998;208:128–134. doi: 10.1007/s004270050163. [DOI] [PubMed] [Google Scholar]
- 30.Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. U. S. A. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsusue K, et al. Liver-specific disruption of PPARγ in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J. Clin. Invest. 2003;111:737–747. doi:10.1172/JCI200317223. doi: 10.1172/JCI17223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zisman A, et al. Targeted disruption of the glucose transporter 4 selectively in muscle causes insulin resistance and glucose intolerance. Nat. Med. 2000;6:924–928. doi: 10.1038/78693. [DOI] [PubMed] [Google Scholar]
- 33.Dudzinski KM, Neff NA. A technique for the combination of clearing, staining, and injecting small mammals. Stain Technol. 1990;65:113–118. doi: 10.3109/10520299009139923. [DOI] [PubMed] [Google Scholar]
- 34.Morgan JE. Myogenicity in vitro and in vivo of mouse muscle cells separated on discontinuous Percoll gradients. J. Neurol. Sci. 1988;85:197–207. doi: 10.1016/0022-510x(88)90156-6. [DOI] [PubMed] [Google Scholar]
- 35.Inscho EW, Wilfinger WW, Banks RO. Dissociation and enrichment of rat atrial myocytes containing atrial natriuretic factor (ANF) Horm. Res. 1986;24:26–37. doi: 10.1159/000180536. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, et al. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat. Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 37.Lapsys NM, et al. Expression of genes involved in lipid metabolism correlate with peroxisome proliferator-activated receptor gamma expression in human skeletal muscle. J. Clin. Endocrinol. Metab. 2000;85:4293–4297. doi: 10.1210/jcem.85.11.6973. [DOI] [PubMed] [Google Scholar]
- 38.Barak Y, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 39.Yamauchi T, et al. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor gamma (PPARgamma) deficiency and PPARgamma agonist improve insulin resistance. J. Biol. Chem. 2001;276:41245–41254. doi: 10.1074/jbc.M103241200. [DOI] [PubMed] [Google Scholar]
- 40.Miles PDG, Barak Y, He W, Evans RM, Olefsky JM. Improved insulin-sensitivity in mice heterozygous for PPAR-gamma deficiency. J. Clin. Invest. 2000;105:287–292. doi: 10.1172/JCI8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ristow M, Muller-Wieland D, Pfeiffer A, Krone W, Kahn CR. Obesity associated with a mutation in a genetic regulator of adipocyte differentiation. N. Engl. J. Med. 1998;339:953–959. doi: 10.1056/NEJM199810013391403. [DOI] [PubMed] [Google Scholar]
- 42.Barroso I, et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- 43.Altshuler D, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 2000;2000:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 44.Deeb SS, et al. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat. Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 45.Kim JK, et al. Glucose toxicity and the development of diabetes in mice with muscle-specific inactivation of GLUT4. J. Clin. Invest. 2001;108:153–160. doi:10.1172/JCI200110294. doi: 10.1172/JCI10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheung AT, Wang J, Ree D, Kolls JK, Bryer-Ash M. Tumor necrosis factor-alpha induces hepatic insulin resistance in obese Zucker (fa/fa) rats via interaction of leukocyte antigen-related tyrosine phosphatase with focal adhesion kinase. Diabetes. 2000;49:810–819. doi: 10.2337/diabetes.49.5.810. [DOI] [PubMed] [Google Scholar]
- 47.Berg AH, Combs TP, Du X, Brownlee M, Scherer PE. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat. Med. 2001;7:947–953. doi: 10.1038/90992. [DOI] [PubMed] [Google Scholar]
- 48.Kelley DE, Mandarino LJ. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes. 2000;49:677–683. doi: 10.2337/diabetes.49.5.677. [DOI] [PubMed] [Google Scholar]
- 49.Ye JM, et al. Peroxisome proliferator-activated receptor (PPAR)-alpha activation lowers muscle lipids and improves insulin sensitivity in high fat-fed rats: comparison with PPAR-gamma activation. Diabetes. 2001;50:411–417. doi: 10.2337/diabetes.50.2.411. [DOI] [PubMed] [Google Scholar]
- 50.Cha BS, et al. Peroxisome proliferator-activated receptor (PPAR) gamma and retinoid X receptor (RXR) agonists have complementary effects on glucose and lipid metabolism in human skeletal muscle. Diabetologia. 2001;44:444–452. doi: 10.1007/s001250051642. [DOI] [PubMed] [Google Scholar]
- 51.Way JM, et al. Comprehensive messenger ribonucleic acid profiling reveals that peroxisome proliferator-activated receptor γ activation has coordinate effects on gene expression in multiple insulin-sensitive tissues. Endocrinology. 2001;142:1269–1277. doi: 10.1210/endo.142.3.8037. [DOI] [PubMed] [Google Scholar]
- 52.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aoyama T, et al. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha) J. Biol. Chem. 1998;273:5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 54.Auboeuf D, et al. Tissue distribution and quantification of the expression of mRNAs of peroxisome proliferator-activated receptors and liver X receptor-alpha in humans: no alteration in adipose tissue of obese and NIDDM patients. Diabetes. 1997;46:1319–1327. doi: 10.2337/diab.46.8.1319. [DOI] [PubMed] [Google Scholar]
- 55.Muoio DM, et al. Fatty acid homeostasis and induction of lipid regulatory genes in skeletal muscles of peroxisome proliferator-activated receptor (PPAR) alpha knock-out mice. Evidence for compensatory regulation by PPAR delta. J. Biol. Chem. 2002;277:26089–26097. doi: 10.1074/jbc.M203997200. [DOI] [PubMed] [Google Scholar]
- 56.Dobbins RL, et al. Prolonged inhibition of muscle carnitine palmitoyltransferase-1 promotes intramyocellular lipid accumulation and insulin resistance in rats. Diabetes. 2001;50:123–130. doi: 10.2337/diabetes.50.1.123. [DOI] [PubMed] [Google Scholar]
- 57.Blaak EE, Aggel-Leijssen DP, Wagenmakers AJ, Saris WH, van Baak MA. Impaired oxidation of plasma-derived fatty acids in type 2 diabetic subjects during moderate-intensity exercise. Diabetes. 2000;49:2102–2107. doi: 10.2337/diabetes.49.12.2102. [DOI] [PubMed] [Google Scholar]
- 58.Blaak EE, et al. Plasma FFA utilization and fatty acid-binding protein content are diminished in type 2 diabetic muscle. Am. J. Physiol. Endocrinol. Metab. 2000;279:E146–E154. doi: 10.1152/ajpendo.2000.279.1.E146. [DOI] [PubMed] [Google Scholar]
- 59.Steppan CM, Lazar MA. Resistin and obesity-associated insulin resistance. Trends Endocrinol. Metab. 2002;13:18–23. doi: 10.1016/s1043-2760(01)00522-7. [DOI] [PubMed] [Google Scholar]
- 60.Hevener AL, Reichart D, Janez A, Olefsky J. Thiazolidinedione treatment prevents free fatty acid-induced insulin resistance in male Wistar rats. Diabetes. 2001;50:2316–2322. doi: 10.2337/diabetes.50.10.2316. [DOI] [PubMed] [Google Scholar]
- 61.McTernan PG, et al. Insulin and rosiglitazone regulation of lipolysis and lipogenesis in human adipose tissue in vitro. Diabetes. 2002;51:1493–1498. doi: 10.2337/diabetes.51.5.1493. [DOI] [PubMed] [Google Scholar]
- 62.Steppan CM, et al. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 63.Maeda N, et al. PPARgamma ligands increase expression and plasma concentrations of adiponectin, an adipose-derived protein. Diabetes. 2001;50:2094–2099. doi: 10.2337/diabetes.50.9.2094. [DOI] [PubMed] [Google Scholar]
- 64.Okuno A, et al. Troglitazone increases the number of small adipocytes without the change of white adipose tissue mass in obese Zucker rats. J. Clin. Invest. 1998;101:1354–1361. doi: 10.1172/JCI1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin G, et al. Induction of the fatty acid transport protein 1 and acyl-CoA synthase genes by dimer-selective rexinoids suggests that the peroxisome proliferator-activated receptor-retinoid X receptor heterodimer is their molecular target. J. Biol. Chem. 2000;275:12612–12618. doi: 10.1074/jbc.275.17.12612. [DOI] [PubMed] [Google Scholar]