Abstract
In the human genome, the majority of protein-encoding genes are interrupted by introns, which are removed from primary transcripts by a macromolecular enzyme known as the spliceosome. Spliceosomes can constitutively remove all the introns in a primary transcript to yield a fully spliced mRNA or alternatively splice primary transcripts leading to the production of many different mRNAs from one gene. This review examines how spliceosomes can recombine two primary transcripts in trans to reprogram messenger RNAs.
Reprogramming of mRNA
The reprogramming of mRNA is a form of gene therapy that modifies mRNA without directly changing the sequence or the transcriptional regulation of the gene. There are two general ways to accomplish mRNA reprogramming. The first alters the processing of primary transcripts without introducing new coding information into these transcripts. An example of this is the inhibition of cryptic or alternative pre-mRNA splicing by antisense oligonucleotides (1), a method that will not be reviewed in this article (see recent review by Kole and Sazani, ref. 2). The second form of mRNA reprogramming involves the recombination of two RNA molecules in trans (see reviews by Sullenger and Gilboa, ref. 3, and Garcia-Blanco et al., ref. 4). Two distinct methods have been used to carry out mRNA reprogramming mediated by trans-splicing. Recombination in trans was first achieved by Sullenger and Cech using a Group I ribozyme designed to bind and trans-splice to a target RNA (5). Using this methodology, several groups have successfully modified the coding potential for mRNAs in prokaryotic and eukaryotic cells (5–8). The second method, which was first described by Puttaraju et al. (9), uses spliceosomes to carry out RNA recombination in trans. Spliceosome-mediated RNA trans-splicing (SMaRT) has been used to reprogram mRNAs in animal cells in culture, in xenografts, and in animals (9–13). SMaRT, and the scientific principles that support it, is the focus of this Perspective series article. A full understanding of this method, its potential, and its limitations requires a discussion of human genes and gene products.
Complex genes and gene products
The sequencing and preliminary annotation of the human genome has provided us with evidence for a preponderance of complex genes and gene products. Transcription of the average human protein-coding gene produces a primary transcript (or pre-mRNA) that spans 27,000–28,000 nucleotides (14, 15). The transcript is composed of introns, usually seven or eight, which are removed during maturation of the primary transcript to mRNA; and exons, usually eight or nine, which are kept in the mRNA (Figure 1). Whereas an average intron is over 3,000 nucleotides in length, the mean size of an exon is 300 nucleotides, and internal exons are a mere 145 nucleotides on average. Thus, the structure of primary transcripts can be viewed as short islands of coding information (exons) surrounded by vast oceans of intronic sequences (Figure 1). The junctions between the short exons and the vast introns, the splice sites, are recognized by spliceosomes. Spliceosomes are macromolecular enzymes that recognize the splice sites and catalyze the removal of introns (16, 17). These large enzymes are composed of five small nuclear uridine-rich small nuclear RNAs and more than sixty proteins, and are very likely ribozymes at their catalytic core. Given the abundance of spliceosomal components it can be assumed that there are between 100,000 and 200,000 spliceosomes per mammalian cell nucleus (ref. 16 and references therein). Spliceosomes must be able to recognize a large number of splice sites. A quick (and likely conservative) calculation suggests that if 10% of all intron-containing genes (∼30,000 genes) are actively transcribing (∼3,000 genes), and each on average is producing three transcripts, then there will be at least 70,000 introns in need of splicing (14, 15). This number, which is of the same order of magnitude as the expected number of spliceosomes per nucleus, suggests that the splicing machinery is efficient and robust. This conclusion has important implications for the development of SMaRT to efficiently reprogram mRNA.
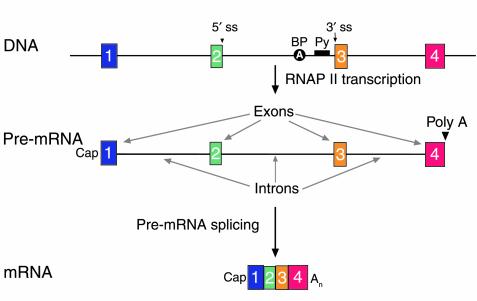
Figure 1.
The structure of human genes and primary transcripts. A schematic of a gene (DNA), its primary transcript (pre-mRNA), and the mature mRNA product. The gene in this schematic spans four exons (boxes 1 through 4) and three introns. Genomic landmarks are indicated for the middle intron: the 5′ splice site (5′ ss), the branchpoint (BP) adenosine, the polypyrimidine tract (Py), and the 3′ splice site (3′ ss). The structure of the pre-mRNA, which is synthesized by RNA polymerase II, is shown. The 5′ end of the pre-mRNA is determined by transcription initiation and modified by a cap structure (27). The 3′ end is created and modified by cleavage and polyadenylation (poly A). Introns are removed in pre-mRNA splicing to yield an mRNA. On average, introns are longer than 3,000 nucleotides. The 5′ and 3′ terminal exons (1 and 4) average 300 nucleotides in length; internal exons (2 and 3) average 145 nucleotides in length (14, 15).
Introns in pre-mRNAs are removed from primary transcripts via two phosphoryl-transfer reactions also known as step 1 and step 2 (Figure 2) (16). These reactions, catalyzed by the spliceosome, must excise the introns precisely in order to produce functional mRNAs. The selection of the splice sites by the spliceosome is guided by sequence elements that define 5′ splice sites and 3′ splice sites (Figure 3). The consensus sequence for 5′ splice sites of the major class of pre-mRNA introns spans an eight-nucleotide sequence (AG|GURAGU, where “|” is the splice site) (14, 17). The consensus for the 3′ splice site spans three elements: the branchpoint sequence, the polypyrimidine tract, and the four-nucleotide sequence at the 3′ splice site (YAG|G) (17). Even though these sequence elements conform to a consensus, there is significant variability (17). The capacity of the spliceosome to recognize variant splice sites underscores the versatility of this enzyme and suggests that SMaRT could be applied to many and perhaps all intron-containing pre-mRNAs (see Figure 3 legend).
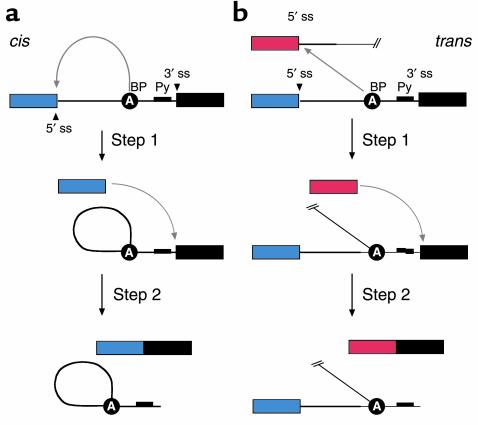
Figure 2.
Both cis-splicing and trans-splicing reactions proceed via two phosphoryl transfer reactions. (a) The cis reaction. A schematic of the two phosphoryl transfer reactions required for intron removal (16). The exons are indicated as boxes (the first or 5′ exon is gray and the second or 3′ exon is black), and the intron is depicted as a line. In the first reaction (step 1), the 2′ OH group of a bulged adenosine at the branchpoint attacks the 5′ phosphate of the first residue of the intron forming the lariat intermediate and the “free” 5′ exon. The lariat contains the branched adenosine, so called because it is connected via conventional 5′ and 3′ links, but also contains a 2′-5′ linkage to the first residue of the intron (see ref. 16 for a more extensive discussion of the splicing reactions). In the second step, the 3′ OH group of the last residue of the “free” 5′ exon attacks the 5′ phosphate of the first residue of the second exon, forming a product with the two exons ligated and releasing the intron as a lariat. (b) The trans reaction. A schematic of the two phosphoryl transfer reactions required for SMaRT. Icons are as described above, except that the trans-splicing or invading exon is shown as a red box. In step 1, the 2′ OH group of the bulged or branchpoint adenosine is again the nucleophile and is attacking the 5′ phosphate of the first residue of an intron in a second RNA molecule (this molecule could be identical in sequence to the first or it could be a completely different RNA). Because the reaction proceeds in trans the branched molecule is now a Y-shaped molecule, not a lariat. Step 2 proceeds as described for step 2 in the cis reaction; however, the exon product includes sequences from two RNAs.
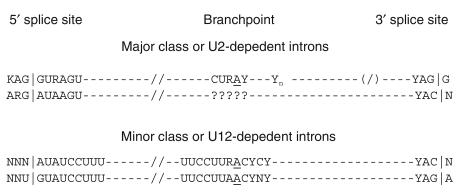
Figure 3.
Consensus sequences at and around the splice sites in nuclear pre-mRNA introns of mammals (17). Two types of introns, U2-dependent and U12-dependent, are spliced by two spliceosomes that share some components. The U2-dependent introns are by far the more prevalent. Although all the published SMaRT reactions to date have targeted U2-dependent introns, there is reason to believe that U12-dependent introns, which represent 0.1% of introns (14), will also be available for targeted SMaRT.
An unforeseen lesson from the sequencing of the human genome was the realization that the majority of primary transcripts are alternatively spliced (Figure 4) (14, 15, 18). A survey of transcripts derived from 245 genes in chromosome 22 found that 145 genes (59%) encoded alternatively spliced transcripts (14). Alternative splicing generates different mRNAs from one gene, leading to the production of proteins with diverse and even antagonistic functions. The example in Figure 4 shows a hypothetical gene with four exons (labeled 1–4) leading to the production of two mRNAs (1•2•4 or 1•3•4) by the exclusive use of exons 2 and 3. The implications of alternative splicing and of its high frequency in the human genome are far-reaching. The dogma “one gene, one polypeptide chain” is not merely a rule with exceptions — in the human genome, it is the exception to the rule. The widespread use of alternative splicing suggests that spliceosomes are versatile enzymes capable of removing a wide variety of introns and ligating diverse exons, and argues against a narrow view of what constitutes a gene and its coding plasticity. Reprogramming of genetic information by sorting exons during alternative splicing is conceptually very similar to reprogramming of mRNAs by spliceosome-mediated trans-splicing. Rather than fooling nature (19), SMaRT follows nature.
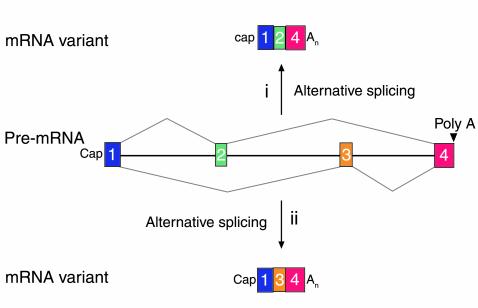
Figure 4.
A schematic of two mRNA variants derived from alternative splicing of a primary transcript. In situation i, exon 2 is recognized whereas exon 3 is not and is read as being part of a large intron. In contrast, the reverse is true for situation ii. In many instances, alternative splicing is tightly regulated in cell type–specific fashion (27). An, refers to the polyA tail at the 3′ end of mRNAs.
Cis-splicing versus trans-splicing
The splicing reactions described above most commonly involve splice sites located within one RNA molecule and are thus considered to occur in cis (Figure 2). In trypanosomes, flatworms, and nematodes, however, specialized spliceosomes can mediate the trans-splicing between highly structured Short Leader RNAs and many different pre-mRNAs (see review and discussion in ref. 20). These specialized trans-splicing reactions are very efficient. In mammals, however, trans-splicing of conventional pre-mRNAs appears to be exceedingly rare, suggesting a mechanistic barrier to trans-splicing (21–24). In principle, the low level of naturally occurring trans-splicing could be due to the presence of trans-acting inhibitors or to the lack of specific trans-activators in mammalian cells. These hypotheses were ruled out when it was shown that specialized Short Leader RNAs could trans-splice to pre-mRNAs in mammalian cells (25). A block of efficient trans-splicing would also be expected if spliceosomes had to scan conventional introns in order to achieve splice site juxtaposition and subsequent splicing. Scanning was essentially ruled out when pre-mRNAs that included obstacles to scanning were shown to splice in vitro with unaltered efficiency (26). A variation of the scanning objection remained plausible, however, since in vivo splicing occurs cotranscriptionally and the elongating RNA polymerase II is likely tethered to spliceosome components that recognize the splice sites (reviewed in refs. 27, 28). This tethering of the splice sites can significantly reduce the likelihood that splice sites in two nascent transcripts will meet. This potential barrier for naturally occurring trans-splicing has to be carefully considered when designing a targeted SMaRT effector.
Reprogramming of mRNA by SMaRT
Targeted spliceosome-mediated RNA trans-splicing requires three components: the spliceosome, a target pre-mRNA, and a SMaRT effector or pre–trans-splicing molecule (PTM) (Figure 5). The spliceosome and the target pre-mRNA are provided by the cell; the PTM is an RNA molecule that is introduced artificially and will be described in detail below. The use of the spliceosome, an efficient and versatile endogenous enzyme, provides a significant advantage over trans-splicing methods that require the introduction of an exogenous ribozyme (5–8). This difference may explain the apparent higher efficiency of spliceosome-mediated mRNA repair (see discussion in ref. 12). The second component, the target pre-mRNA, is the precursor to the mRNA to be reprogrammed (Figure 5). In order to be a substrate for spliceosome-mediated trans-splicing, this target pre-mRNA must contain at least one intron, and the reprogramming can be achieved only by trans-splicing at the naturally occurring splice sites in the target although the requirement for naturally occurring splice sites represents a limitation relative to Group I ribozyme–mediated targeted trans-splicing, the number of gene targets amenable to reprogramming by targeted SMaRT is likely to exceed 90% of protein-encoding genes (14, 15). Dependence on expression of the pre-mRNA target can provide trans-splicing with cell-type specificity, which can be further enhanced by regulating the expression pattern of the PTM. This advantage of target dependence is shared by all trans-splicing methodologies over techniques that use direct expression of genes. The two cellular components used by SMaRT, the spliceosome and the target pre-mRNA, provide these trans-splicing reactions with unique advantages over conventional gene therapy strategies.
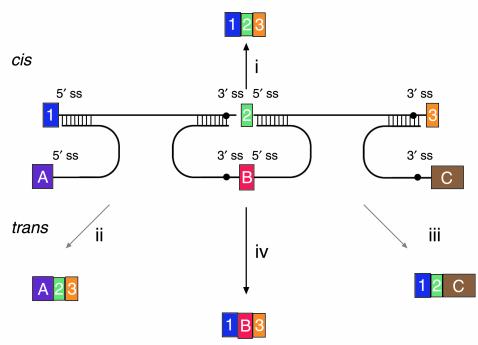
Figure 5.
Versatility of mRNA reprogramming by targeted SMaRT. The figure shows a schematic of splicing reactions involving a three-exon pre-mRNA and PTMs. A conventional constitutive cis-splicing reaction leading to the production of the expected 1•2•3 mRNA is shown (i). Three targeted SMaRT reactions are shown. PTM[A] contains a functional 5′ splice site that can trans-splice to the 3′ splice site adjacent to exon 2 in the pre-mRNA target (ii). This trans-splicing produces a chimeric A•2•3 mRNA. PTM[A] is targeted to occlude the naturally occurring 5′ splice site at exon 1 to reduce the use of this site. PTM[C] contains a functional 3′ splice site that can trans-splice to the 5′ splice site adjacent to exon 2 in the pre-mRNA target (iii). This trans-splicing produces a chimeric 1•2•C mRNA. Finally, PTM[B] contains both 3′ and 5′ splice sites bordering an exon, and these splice sites can trans-splice with the 5′ splice site adjacent to exon 1 and the 5′ splice site adjacent to exon 3, respectively (iv). These two trans-splicing reactions lead to internal exon replacement and produce the chimeric 1•B•3 mRNA.
The third component in the spliceosome-mediated trans-splicing reactions is the PTM. PTMs can be designed to carry out one of three forms of trans-splicing, depending on the type of trans-splicing domain in the PTM (Figure 5) (see discussions in refs. 4, 9). These domains provide PTMs with the cis elements required for spliceosome recognition and splicing. A PTM with a 5′ splice site domain can trans-splice to a 3′ splice site in the target pre-mRNA (ii in Figure 5); a PTM with a 3′ splice site domain can trans-splice to a 5′ splice site in the target (iii); and finally, a PTM with dual 3′ and 5′ splice site domains can precisely replace an internal exon (iv). In addition to the trans-splicing domain, PTMs include a binding domain responsible for specific targeting of the PTM and a coding domain that includes the new or modified genetic information that will reprogram the target (Figure 6). A more extensive discussion of PTM structure by Garcia-Blanco et al. can be reviewed in ref. 4. It must be noted that PTMs are modular and versatile, permitting the independent development of the trans-splicing, binding, and coding domains. For instance, PTM[C] in Figure 5 could be modified to include the binding domain of PTM[B]. This new PTM would be directed to splice with exon 1 in the target pre-mRNA, and the trans-splicing would yield a 1•C mRNA rather than the 1•2•C mRNA. The length and composition of the binding domains can be modified to alter efficiency, specificity, and targeting location within a pre-mRNA (12). Equally, the coding domain can be modified to enhance protein expression and function. The open reading frame can be modified to maximize translation (e.g., optimizing codon use to match preferences) and to improve protein function (e.g., introduction of up mutations). Expression can also be regulated or enhanced by alterations of the untranslated regions of the coding domain (see Figure 6). In addition to carrying target-binding, trans-splicing, and coding domains, the PTMs or the DNA molecules that encode them must carry instructions to deliver the PTMs to their targets.
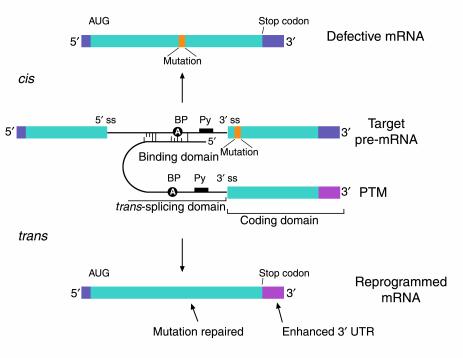
Figure 6.
The anatomy of a PTM. The figure depicts a mutant target pre-mRNA (see the LacZ model system in ref. 12) that is incapable of coding for functional protein because it contains a mutation in the second exon (indicated as orange segment). The product of the conventional cis-splicing reaction for this pre-mRNA is a defective mRNA. The PTM shown contains several domains. The binding domain (also referred to as the targeting domain) binds to the pre-mRNA target, thus localizing the PTM near the site of the desired trans-splicing reaction. The binding domain can also be designed to occlude important elements within the 3′ splice site of the target and thus reduce the cis-splicing reaction. Between the binding domain and the trans-splicing domain there is usually a spacer region for flexibility. The trans-splicing domain, which in the case shown includes the elements required to make a potent 3′ splice site, is responsible for the reactivity of the PTM. Finally, the coding domain contains the necessary genetic information that will be imparted into the reprogrammed RNA. This can include protein-coding instructions as well as instructions for the effective processing, transport, and localization of the reprogrammed mRNA. In the example shown, the open reading frame has been changed to repair the mutation in the gene, and the 3′ untranslated region (3′ UTR) has been enhanced to increase mRNA stability and translation. It should be noted that the cis- and trans-splicing reactions are in competition. This implies that as the level of reprogrammed mRNA increases, the level of defective mRNA decreases. A decrease in the level of defective mRNA may be very useful in the case of dominant mutations.
PTMs must be delivered into the nucleus of cells to find the targeted pre-mRNAs. PTMs can either be introduced directly into the nucleus or synthesized in the nucleus from DNA templates. Synthesis from DNA templates has been most successful to date (9–13). PTM templates must be delivered to the nucleus, a requirement that most gene therapeutics share. Beyond nuclear delivery, effective PTM trans-splicing depends on finding the target pre-mRNA as a nascent transcript (27). Therefore, the ideal PTM template would home in to the transcriptional domain of the target pre-mRNA, synthesizing the PTM at or very near the site of formation of the target pre-mRNA. Although this idealized type of nuclear localization has yet to be accomplished, the targeting of PTMs in living cells has been studied using tissue culture systems (9–13).
SMaRT in living cells
Targeted SMaRT was first achieved in vitro in the mid-1980s (29, 30). These early experiments suggested that tethering the two partners in trans-splicing reactions would greatly enhance the rate of splicing. The trans-splicing reactions were proposed to follow a similar mechanism to that observed in conventional cis-splicing, i.e., two phosphoryl transfer reactions (Figure 2). Although these studies showed that spliceosomes could direct targeted trans-splicing in vitro, the question remained whether it would be possible to use SMaRT to reprogram a conventional human mRNA in a living cell. In 1999, Puttaraju et al. showed for the first time that a PTM could be used to reprogram the human chorionic gonadotropin β-polypeptide (CGB) mRNA in human H1299 tumor cells (9). The trans-splicing reaction was precise, producing a chimeric messenger between the CGB transcript and the coding domain of a PTM, which encoded the A subunit of diphtheria toxin (DT-A). Evidence that the chimeric CGB•DT-A mRNAs were functional came from toxicity studies (4). Direct evidence for functional mRNA reconstruction was obtained using HEK 293 cells engineered to harbor a defective LacZ gene. This defective LacZ gene was a target for SMaRT because it was split by an intron derived from the human cystic fibrosis transmembrane conductance regulator (CFTR) gene (9, 12) (Figure 5). The defect in the LacZ gene was engineered by including two in-frame stop codons in the second exon, which rendered the gene incapable of directing the synthesis of β-galactosidase (Figure 6). Not only was the defect in the LacZ mRNA repaired by SMaRT, but the cells transfected with the appropriate PTM produced full-length β-galactosidase protein and had β-galactosidase activity (12). Although this was the first report of restoration of endogenous protein structure and function by trans-splicing in mammalian cells, it was nonetheless reprogramming of an artificial gene with a very simple structure. To test the utility of SMaRT in reprogramming a human gene with clinical relevance, PTMs were developed to repair CFTR mRNA (10). Recent and very encouraging results show that PTMs targeted to the endogenous Δ508 CFTR pre-mRNA lead to mRNA repair and partial restoration of Cl– transport defects in human CF airway epithelial cells in culture (13). Genes encoding the corrective PTMs were delivered using adenoviral vectors. At a multiplicity of infection of 2,000 viral particles/cell, the Cl– current was found to be between 12% and 16% of that observed in non-CF epithelial cultures (13). This level of correction would be enough for a therapeutic response (13, 19). These studies clearly demonstrate that SMaRT can be used to reprogram mRNAs and restore or alter protein structure and function in living cells.
The studies in tissue culture systems also highlight the importance of two critical issues: efficiency and specificity (see also discussion in ref. 19). How well does SMaRT work in living cells? In experiments that targeted the LacZ pre-mRNA in HEK 293 cells, trans-splicing efficiency was measured at 2% of LacZ mRNA levels (12). This measurement was carried out using a real-time quantitative RT-PCR method developed by Baker and colleagues (12). The level of protein repair has been harder to assess, but levels below 1% were estimated for CFTR protein correction (see discussion in ref. 13). Although these numbers for mRNA and protein correction are low, the effect on enzymatic restoration for CFTR was significantly higher (12–16%) (13). Although the apparent disparity between protein levels and CFTR-dependent Cl– transport may indicate that only activity measurements are relevant in determining efficacy, more cases need to be analyzed carefully to truly quantify the effectiveness of SMaRT. These prior considerations notwithstanding, the efficiency of PTMs should be increased considerably in order to expand the utility of these reagents. To date, the most significant improvements in activity have been made by increasing the length of the binding domain (12, 13). Short binding domains, which may be optimal for stem formation in vitro, probably form unstable stems in vivo due to nuclear helicases (17), whereas long binding domains presumably stabilize the PTM-target interaction. A rigorous study of the effect of length and composition of the binding domain on efficiency should be carried out. To increase trans-splicing efficiency, PTMs could be modified by including splicing enhancer elements, preferably those found in introns (30). Additionally, targeting of 3′ splice site PTMs to a site near the pre-mRNA 5′ splice site would take advantage of the kinetics of cotranscriptional splicing and possibly enhance efficiency (27).
A second critical issue that can be addressed in tissue culture model systems is that of specificity. Synthesis of nonspecific products after introduction of PTM into cells can be mediated by inappropriate direct expression of PTM-encoded products or by nontargeted trans-splicing to an irrelevant cellular pre-mRNA or to inappropriate splice sites within the targeted pre-mRNA. All of these types of products have been observed in cells expressing PTMs (10–12). To decrease the direct synthesis of undesired products, PTMs have been modified to diminish spurious translation initiation (10, 12). To prevent nonspecific trans-splicing to pre-mRNAs, several modifications have been designed. PTMs were designed so that the splice sites were occluded by a stem structure until the binding domain was engaged on the target pre-mRNA. These PTMs successfully diminished nonspecific trans-splicing in vitro and in vivo in some cases (9, 10) but not in others (11). Increasing the size of binding domains to more than 200 nucleotides dramatically reduced the ratio of nonspecific to specific trans-splicing in the LacZ model system (12). Comparisons between studies have not been easy because different methods have been used to measure specificity the trans-splicing reactions (11, 12). Clear improvements in specificity will be required to prevent the formation of unwanted chimeric proteins, some of which could be toxic and/or immunogenic.
Even though these tissue culture studies reveal many unresolved problems, it is apparent that SMaRT can repair mRNAs and restore proteins and their activities in mammalian cells. The restoration of function in mammalian cells in culture is encouraging and has provided the impetus for the exploration of SMaRT in animal models of human disease.
Experiments in animal model systems
A critical step in the evaluation of any new therapeutic is its ability to perform in animal models. Such studies have recently begun for SMaRT, and although it is quite early, some reports are worth summarizing. The first demonstration of SMaRT in animals was the targeting of endogenous CGB transcripts in H1299 tumor xenografts in nude mice (9). Although very low levels of trans-splicing were obtained, this experiment showed that SMaRT could be achieved in an animal. More recently, two exciting developments have been reported. SMaRT was used to partially correct a Δ508 CFTR defect in human bronchial CF xenografts in nude mice (13). The level of Cl– permeability obtained with SMaRT was consistent with CFTR activation at 22% of that observed in normal bronchial xenografts (13). SMaRT has also been used to partially restore a Factor VIII defect in a hemophilia A mouse model system (31). Levels of circulating Factor VIII were increased from essentially undetectable levels to 12% of the levels found in normal mice. The levels of CFTR and Factor VIII restoration in these animals are sufficient for a significant therapeutic effect (13). Both of these studies, however, analyzed mRNA reprogramming in transient assays, leaving unanswered questions about the persistence of SMaRT repair. Nonetheless, these studies represent a promising beginning and should be rapidly followed by assessments of long-term efficiency, specificity, and safety in these and other animal model systems.
Although this Perspective series article will not focus on the issue of delivery, it should be noted that trans-splicing can lead to several advantages over conventional gene therapy. First, trans-splicing technologies do not require that the complete gene sequences be introduced. In fact, internal exon replacement reactions can conceivably repair a mutated exon with delivery of PTMs shorter than 250 nucleotides. The smaller genetic package required provides two advantages. Firstly, the choice of delivery vector (viral or nonviral) is wider, and secondly, there is more room to add regulatory elements to control the cell type–specific expression of the PTM. This is particularly important when using vectors with very limited packaging size (e.g., adeno-associated virus) (see discussion in refs. 13, 19). Another advantage of trans-splicing technologies in terms of delivery is the ability to multiply the specificity obtained by controlling expression of the PTM with the specificity provided by cell type–specific target expression. This releases constraints for strict specificity of delivery. As is true of all other gene therapy modalities, overall efficacy of SMaRT will be significantly influenced by the efficiency, specificity, and safety of gene delivery systems.
The early animal studies summarized above suggest promise for the therapeutic application of SMaRT to many human genetic disorders, possibly even dominant mutations. Much work still needs to be done both in these animal model systems and in the basic scientific design of the PTMs. The goal should be to enhance, by at least an order of magnitude, the efficiency and the specificity of PTMs. The current state of the art merits cautious optimism for the therapeutic potential of SMaRT. In addition, nontherapeutic applications of SMaRT, which fall outside the scope of this article, also hold great promise (4).
Acknowledgments
M.A. Garcia-Blanco wishes to thank Glen Coburn, Gerard McGarrity, and Christopher Walsh for critical reading of this manuscript, and Annette Kennett for her assistance in the preparation of this manuscript. M.A. Garcia-Blanco was a scholar of the Raymond and Beverly Sackler Foundation, and he also acknowledges funding from the NIH.
Footnotes
Conflict of interest: The author is a scientific founder and consultant of Intronn Inc., which is commercializing therapeutic and nontherapeutic applications of SMaRT.
Nonstandard abbreviations used: spliceosome-mediated RNA trans-splicing (SMaRT); pre–trans-splicing molecule (PTM); chorionic gonadotropin β-polypeptide (CGB); diphtheria toxin A subunit (DT-A); cystic fibrosis transmembrane conductance regulator (CFTR).
References
- 1.Dominski Z, Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kole R, Sazani P. Antisense effects in the nucleus: modification of splicing. Curr. Opin. Mol. Ther. 2001;3:229–234. [PubMed] [Google Scholar]
- 3.Sullenger BA, Gilboa E. Emerging clinical applications of RNA. Nature. 2002;418:252–258. doi: 10.1038/418252a. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Blanco MA, Puttaraju M, Mansfield SG, Mitchell LG. Spliceosome-mediated RNA trans-splicing in gene therapy and genomics. Gene Ther. Reg. 2000;1:141–163. doi: 10.1038/sj.gt.3301307. [DOI] [PubMed] [Google Scholar]
- 5.Sullenger B, Cech TR. Ribozyme-mediated repair of defective mRNA by targeted trans-splicing. Nature. 1994;317:619–622. doi: 10.1038/371619a0. [DOI] [PubMed] [Google Scholar]
- 6.Lan N, Howrey RP, Lee SW, Smith CA, Sullenger B. Ribozyme-mediated repair of sickly β-globin mRNAs in erythrocyte precursors. Science. 1998;280:1593–1596. doi: 10.1126/science.280.5369.1593. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe T, Sullenger B. Induction of wild-type p53 activity in human cancer cells by ribozymes that repair mutant p53 transcripts. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8490–8494. doi: 10.1073/pnas.150104097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayre BG, Kohler U, Goodman HM, Haselhoff J. Design of highly specific cytotoxins by using trans-splicing ribozymes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3507–3512. doi: 10.1073/pnas.96.7.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puttaraju M, Jamison SF, Mansfield SG, Garcia-Blanco MA, Mitchell LG. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotech. 1999;17:246–252. doi: 10.1038/6986. [DOI] [PubMed] [Google Scholar]
- 10.Mansfield SG, et al. Repair of CFTR mRNA by spliceosome-mediated RNA trans-splicing. Gene Ther. 2000;7:1885–1895. doi: 10.1038/sj.gt.3301307. [DOI] [PubMed] [Google Scholar]
- 11.Kikumori T, Cote G, Gagel RF. Promiscuity of pre-mRNA spliceosome-mediated trans-splicing: a problem for gene therapy? Hum. Gene. Ther. 2001;12:1429–1441. doi: 10.1089/104303401750298580. [DOI] [PubMed] [Google Scholar]
- 12.Puttaraju M, DiPasquale J, Baker CC, Mitchell LG, Garcia-Blanco MA. Messenger RNA repair and restoration of protein function by spliceosome-mediated RNA trans-splicing. Mol. Ther. 2001;4:105–114. doi: 10.1006/mthe.2001.0426. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, et al. Partial correction of endogenous ΔF508 CFTR in human cystic fibrosis airway epithelia by spliceosome-mediated RNA trans-splicing. Nat. Biotech. 2002;20:47–52. doi: 10.1038/nbt0102-47. [DOI] [PubMed] [Google Scholar]
- 14.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 15.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Blanco, M.A., Lindsey, L., and Ghosh, S. 2001. The phosphoryl transfer reactions in pre-messenger RNA splicing. In RNA. D. Soll, S. Nishimura, and P. Moore, editors. Elsevier Science/Pergamon. Amsterdam, The Netherlands. 109–124.
- 17.Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosomes. In The RNA world. R.F. Gesteland, T.R. Cech, and J.F. Atkins, editors. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 525–560.
- 18.Claverie JM. Gene number: what if there are only 30,000 human genes? Science. 2001;291:1255–1257. doi: 10.1126/science.1058969. [DOI] [PubMed] [Google Scholar]
- 19.Crystal RC. Fooling mother nature. Nat. Biotech. 2002;20:32–33. doi: 10.1038/nbt0102-32. [DOI] [PubMed] [Google Scholar]
- 20.Nilsen TW. Evolutionary origin of SL-addition trans-splicing: still an enigma. Trends Genet. 2001;17:678–680. doi: 10.1016/s0168-9525(01)02499-4. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan PM, Petrusz P, Szpirer C, Joseph DR. Alternative processing of androgen-binding protein RNA transcripts in fetal rat liver. Identification of a transcript formed by trans splicing. J. Biol. Chem. 1991;266:143–154. [PubMed] [Google Scholar]
- 22.Eul J, Graessmann M, Graessmann A. Experimental evidence for RNA trans-splicing in mammalian cells. EMBO J. 1995;14:3226–3235. doi: 10.1002/j.1460-2075.1995.tb07325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caudevilla C, et al. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12185–12190. doi: 10.1073/pnas.95.21.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tasic B, et al. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell. 2002;10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 25.Bruzik JP, Maniatis T. Spliced leader RNAs from lower eukaryotes are trans-spliced in mammalian cells. Nature. 1992;360:692–695. doi: 10.1038/360692a0. [DOI] [PubMed] [Google Scholar]
- 26.Pasman Z, Garcia-Blanco MA. The 5′ and 3′ splice sites come together via a three dimensional diffusion mechanism. Nucleic Acids Res. 1996;24:1638–1645. doi: 10.1093/nar/24.9.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldstrohm AC, Greenleaf AL, Garcia-Blanco MA. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene. 2001;277:31–47. doi: 10.1016/s0378-1119(01)00695-3. [DOI] [PubMed] [Google Scholar]
- 28.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 29.Konarska MM, Padgett RA, Sharp PA. Trans-splicing of mRNA precursors in vitro. Cell. 1985;42:165–171. doi: 10.1016/s0092-8674(85)80112-4. [DOI] [PubMed] [Google Scholar]
- 30.Solnick D. Trans-splicing of mRNA precursors. Cell. 1985;42:157–164. doi: 10.1016/s0092-8674(85)80111-2. [DOI] [PubMed] [Google Scholar]
- 31.Chao, H., et al. 2003. Phenotype correction of hemophilia A mice by spliceosome-mediated RNA trans-splicing. Nat. Med. In press. [DOI] [PubMed]