Abstract
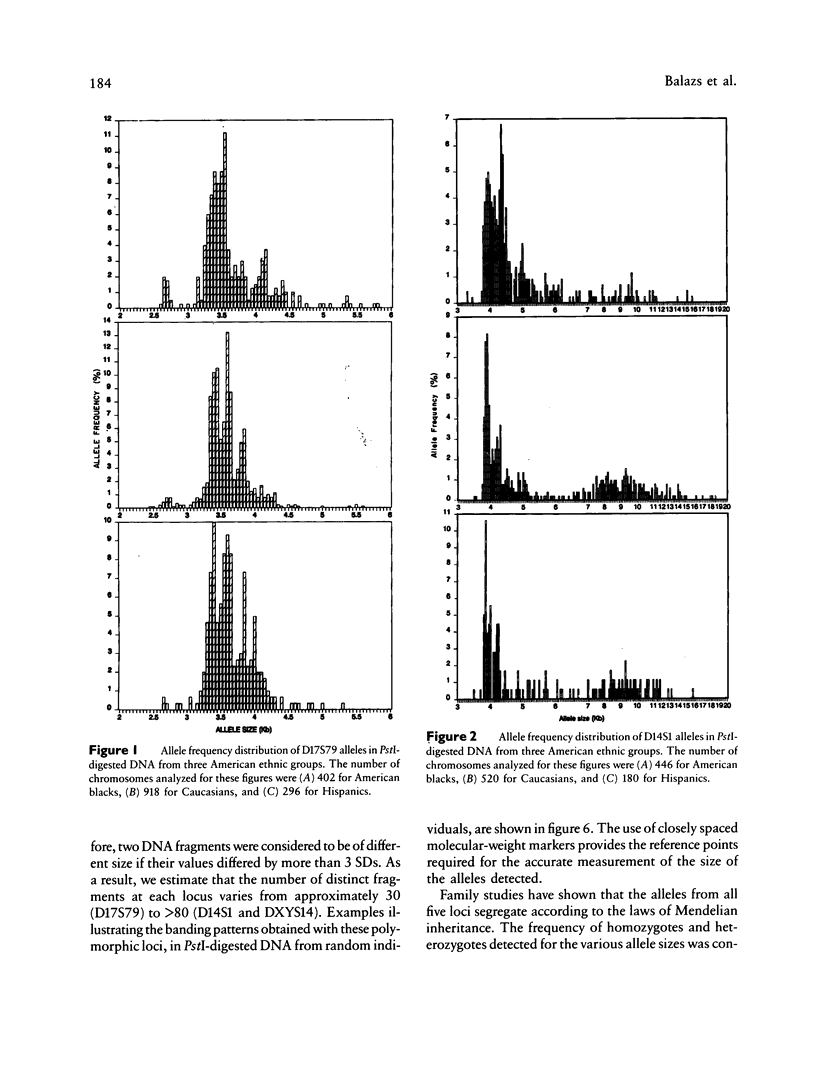
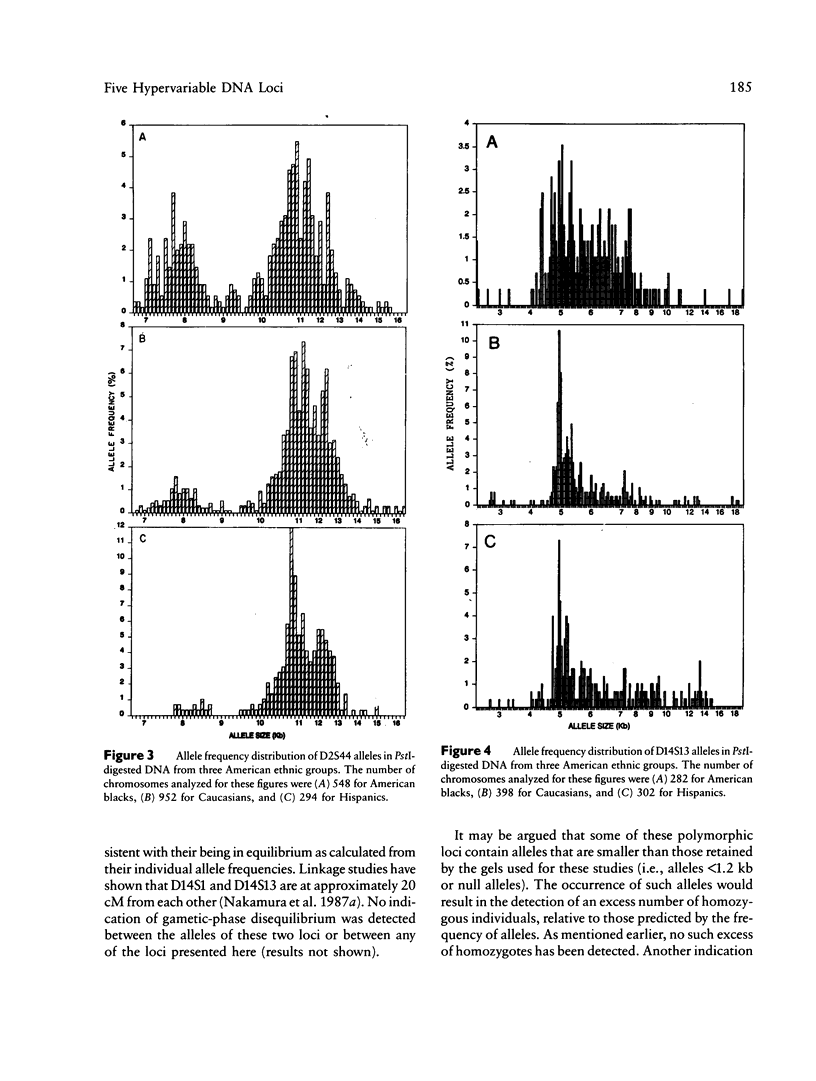
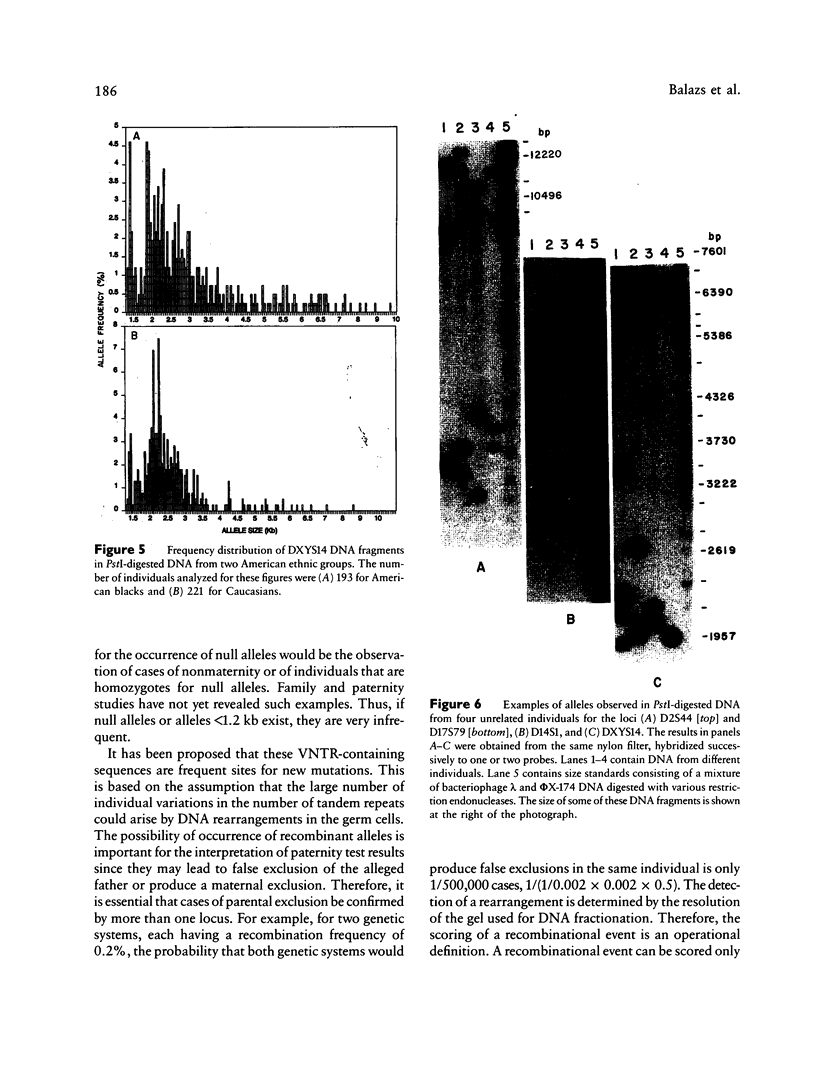
Population genetic studies were performed using DNA probes that recognize five hypervariable loci (D2S44, D14S1, D14S13, D17S79, and DXYS14) in the human genome. DNA from approximately 900 unrelated individuals, subdivided into three ethnic groups (American blacks, Caucasians, and Hispanics) were digested with PstI and were successively hybridized to each DNA probe. The number of distinct DNA fragments identified for each of these regions varies from 30 to more than 80. An allele frequency distribution was determined for each locus and each ethnic group. The results show significant differences, between ethnic groups, in the pattern of distribution as well as in the relative frequency of the most common alleles of D2S44, D14S1, and D14S13 but only small differences in others (i.e., D17S79 and DXYS14). The results presented show that the analysis of these loci can have useful applications in population genetics as well as in identity tests.
Full text
PDF





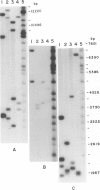
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baird M., Balazs I., Giusti A., Miyazaki L., Nicholas L., Wexler K., Kanter E., Glassberg J., Allen F., Rubinstein P. Allele frequency distribution of two highly polymorphic DNA sequences in three ethnic groups and its application to the determination of paternity. Am J Hum Genet. 1986 Oct;39(4):489–501. [PMC free article] [PubMed] [Google Scholar]
- Bell G. I., Selby M. J., Rutter W. J. The highly polymorphic region near the human insulin gene is composed of simple tandemly repeating sequences. Nature. 1982 Jan 7;295(5844):31–35. doi: 10.1038/295031a0. [DOI] [PubMed] [Google Scholar]
- Capon D. J., Chen E. Y., Levinson A. D., Seeburg P. H., Goeddel D. V. Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature. 1983 Mar 3;302(5903):33–37. doi: 10.1038/302033a0. [DOI] [PubMed] [Google Scholar]
- Cooke H. J., Brown W. R., Rappold G. A. Hypervariable telomeric sequences from the human sex chromosomes are pseudoautosomal. Nature. 1985 Oct 24;317(6039):687–692. doi: 10.1038/317687a0. [DOI] [PubMed] [Google Scholar]
- Cooke H. J., Smith B. A. Variability at the telomeres of the human X/Y pseudoautosomal region. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):213–219. doi: 10.1101/sqb.1986.051.01.026. [DOI] [PubMed] [Google Scholar]
- FISHER R. A. Standard calculations for evaluating a blood-group system. Heredity (Edinb) 1951 Apr;5(1):95–102. doi: 10.1038/hdy.1951.5. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Ginsburg D., Antin J. H., Smith B. R., Orkin S. H., Rappeport J. M. Origin of cell populations after bone marrow transplantation. Analysis using DNA sequence polymorphisms. J Clin Invest. 1985 Feb;75(2):596–603. doi: 10.1172/JCI111736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti A., Baird M., Pasquale S., Balazs I., Glassberg J. Application of deoxyribonucleic acid (DNA) polymorphisms to the analysis of DNA recovered from sperm. J Forensic Sci. 1986 Apr;31(2):409–417. [PubMed] [Google Scholar]
- Ito H., Yasuda N., Matsumoto H. The probability of parentage exclusion based on restriction fragment length polymorphisms. Jinrui Idengaku Zasshi. 1985 Dec;30(4):261–269. doi: 10.1007/BF01907963. [DOI] [PubMed] [Google Scholar]
- Jarman A. P., Nicholls R. D., Weatherall D. J., Clegg J. B., Higgs D. R. Molecular characterisation of a hypervariable region downstream of the human alpha-globin gene cluster. EMBO J. 1986 Aug;5(8):1857–1863. doi: 10.1002/j.1460-2075.1986.tb04437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Royle N. J., Wilson V., Wong Z. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature. 1988 Mar 17;332(6161):278–281. doi: 10.1038/332278a0. [DOI] [PubMed] [Google Scholar]
- Kanter E., Baird M., Shaler R., Balazs I. Analysis of restriction fragment length polymorphisms in deoxyribonucleic acid (DNA) recovered from dried bloodstains. J Forensic Sci. 1986 Apr;31(2):403–408. [PubMed] [Google Scholar]
- Nakamura Y., Leppert M., O'Connell P., Wolff R., Holm T., Culver M., Martin C., Fujimoto E., Hoff M., Kumlin E. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987 Mar 27;235(4796):1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wainscoat J. S., Pilkington S., Peto T. E., Bell J. I., Higgs D. R. Allele-specific DNA identity patterns. Hum Genet. 1987 Apr;75(4):384–387. doi: 10.1007/BF00284114. [DOI] [PubMed] [Google Scholar]