Abstract
A wide spectrum of mutations, ranging from point mutations to large deletions, have been described in the retinoblastoma gene (RB1). Mutations have been found throughout the gene; however, these genetic alterations do not appear to be homogeneously distributed. In particular, a significant proportion of disease-causing mutations results in the premature termination of protein synthesis, and the majority of these mutations occur as C-->T transitions at CpG dinucleotides (CpGs). Such recurrent CpG mutations, including those found in RB1, are likely the result of the deamination of 5-methylcytosine within these CpGs. In the present study, we used the sodiumbisulfite conversion method to detect cytosine methylation in representative exons of RB1. We analyzed DNA from a variety of tissues and specifically targeted CGA codons in RB1, where recurrent premature termination mutations have been reported. We found that DNA methylation within RB1 exons 8, 14, 25, and 27 appeared to be restricted to CpGs, including six CGA codons. Other codons containing methylated cytosines have not been reported to be mutated. Therefore, disease-causing mutations at CpGs in RB1 appear to be determined by several factors, including the constitutive presence of DNA methylation at cytosines within CpGs, the specific codon within which the methylated cytosine is located, and the particular region of the gene within which that codon resides.
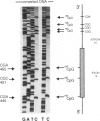
Full text
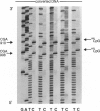
PDF





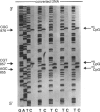
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews J. D., Mancini D. N., Singh S. M., Rodenhiser D. I. Site and sequence specific DNA methylation in the neurofibromatosis (NF1) gene includes C5839T: the site of the recurrent substitution mutation in exon 31. Hum Mol Genet. 1996 Apr;5(4):503–507. doi: 10.1093/hmg/5.4.503. [DOI] [PubMed] [Google Scholar]
- Antequera F., Boyes J., Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990 Aug 10;62(3):503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Barlow D. P. Methylation and imprinting: from host defense to gene regulation? Science. 1993 Apr 16;260(5106):309–310. doi: 10.1126/science.8469984. [DOI] [PubMed] [Google Scholar]
- Blanquet V., Turleau C., Gross-Morand M. S., Sénamaud-Beaufort C., Doz F., Besmond C. Spectrum of germline mutations in the RB1 gene: a study of 232 patients with hereditary and non hereditary retinoblastoma. Hum Mol Genet. 1995 Mar;4(3):383–388. doi: 10.1093/hmg/4.3.383. [DOI] [PubMed] [Google Scholar]
- Canning S., Dryja T. P. Short, direct repeats at the breakpoints of deletions of the retinoblastoma gene. Proc Natl Acad Sci U S A. 1989 Jul;86(13):5044–5048. doi: 10.1073/pnas.86.13.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. J., Harrison J., Frommer M. CpNpG methylation in mammalian cells. Nat Genet. 1995 May;10(1):20–27. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- Clark S. J., Harrison J., Paul C. L., Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994 Aug 11;22(15):2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comb M., Goodman H. M. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990 Jul 11;18(13):3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. N., Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet. 1989 Sep;83(2):181–188. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- Cooper D. N., Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988 Feb;78(2):151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Cowell J. K., Smith T., Bia B. Frequent constitutional C to T mutations in CGA-arginine codons in the RB1 gene produce premature stop codons in patients with bilateral (hereditary) retinoblastoma. Eur J Hum Genet. 1994;2(4):281–290. doi: 10.1159/000472372. [DOI] [PubMed] [Google Scholar]
- Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., Molloy P. L., Paul C. L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger V., Debus N., Lohmann D., Höpping W., Passarge E., Horsthemke B. Frequency and parental origin of hypermethylated RB1 alleles in retinoblastoma. Hum Genet. 1994 Nov;94(5):491–496. doi: 10.1007/BF00211013. [DOI] [PubMed] [Google Scholar]
- Greger V., Passarge E., Höpping W., Messmer E., Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989 Sep;83(2):155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- Henson J. W., Schnitker B. L., Correa K. M., von Deimling A., Fassbender F., Xu H. J., Benedict W. F., Yandell D. W., Louis D. N. The retinoblastoma gene is involved in malignant progression of astrocytomas. Ann Neurol. 1994 Nov;36(5):714–721. doi: 10.1002/ana.410360505. [DOI] [PubMed] [Google Scholar]
- Hogg A., Bia B., Onadim Z., Cowell J. K. Molecular mechanisms of oncogenic mutations in tumors from patients with bilateral and unilateral retinoblastoma. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7351–7355. doi: 10.1073/pnas.90.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höller M., Westin G., Jiricny J., Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988 Sep;2(9):1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- Iguchi-Ariga S. M., Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989 May;3(5):612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- Kato M. V., Ishizaki K., Toguchida J., Kaneko A., Takayama J., Tanooka H., Kato T., Shimizu T., Sasaki M. S. Mutations in the retinoblastoma gene and their expression in somatic and tumor cells of patients with hereditary retinoblastoma. Hum Mutat. 1994;3(1):44–51. doi: 10.1002/humu.1380030108. [DOI] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Laird P. W., Jaenisch R. DNA methylation and cancer. Hum Mol Genet. 1994;3(Spec No):1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- Leteurtre F., Kohlhagen G., Fesen M. R., Tanizawa A., Kohn K. W., Pommier Y. Effects of DNA methylation on topoisomerase I and II cleavage activities. J Biol Chem. 1994 Mar 18;269(11):7893–7900. [PubMed] [Google Scholar]
- Lohmann D. R., Brandt B., Höpping W., Passarge E., Horsthemke B. Spectrum of small length germline mutations in the RB1 gene. Hum Mol Genet. 1994 Dec;3(12):2187–2193. doi: 10.1093/hmg/3.12.2187. [DOI] [PubMed] [Google Scholar]
- Magewu A. N., Jones P. A. Ubiquitous and tenacious methylation of the CpG site in codon 248 of the p53 gene may explain its frequent appearance as a mutational hot spot in human cancer. Mol Cell Biol. 1994 Jun;14(6):4225–4232. doi: 10.1128/mcb.14.6.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur M., Thomas J. O. A preference of histone H1 for methylated DNA. EMBO J. 1996 Apr 1;15(7):1705–1714. [PMC free article] [PubMed] [Google Scholar]
- Nan X., Tate P., Li E., Bird A. DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol. 1996 Jan;16(1):414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory K., Legros Y., Auguin C., Soussi T. Analysis of the most representative tumour-derived p53 mutants reveals that changes in protein conformation are not correlated with loss of transactivation or inhibition of cell proliferation. EMBO J. 1994 Aug 1;13(15):3496–3504. doi: 10.1002/j.1460-2075.1994.tb06656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny I. P., Poirier L. A., James S. J. Differential sensitivity to loss of cytosine methyl groups within the hepatic p53 gene of folate/methyl deficient rats. Carcinogenesis. 1995 Nov;16(11):2863–2867. doi: 10.1093/carcin/16.11.2863. [DOI] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991 Sep;55(3):451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenhiser D. I., Andrews J. D., Mancini D. N., Jung J. H., Singh S. M. Homonucleotide tracts, short repeats and CpG/CpNpG motifs are frequent sites for heterogeneous mutations in the neurofibromatosis type 1 (NF1) tumour-suppressor gene. Mutat Res. 1997 Feb 3;373(2):185–195. doi: 10.1016/s0027-5107(96)00171-6. [DOI] [PubMed] [Google Scholar]
- Rodenhiser D., Chakraborty P., Andrews J., Ainsworth P., Mancini D., Lopes E., Singh S. Heterogenous point mutations in the BRCA1 breast cancer susceptibility gene occur in high frequency at the site of homonucleotide tracts, short repeats and methylatable CpG/CpNpG motifs. Oncogene. 1996 Jun 20;12(12):2623–2629. [PubMed] [Google Scholar]
- Sakai T., Toguchida J., Ohtani N., Yandell D. W., Rapaport J. M., Dryja T. P. Allele-specific hypermethylation of the retinoblastoma tumor-suppressor gene. Am J Hum Genet. 1991 May;48(5):880–888. [PMC free article] [PubMed] [Google Scholar]
- Tate P., Skarnes W., Bird A. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet. 1996 Feb;12(2):205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- Tornaletti S., Pfeifer G. P. Complete and tissue-independent methylation of CpG sites in the p53 gene: implications for mutations in human cancers. Oncogene. 1995 Apr 20;10(8):1493–1499. [PubMed] [Google Scholar]
- Van Orsouw N. J., Li D., van der Vlies P., Scheffer H., Eng C., Buys C. H., Li F. P., Vijg J. Mutational scanning of large genes by extensive PCR multiplexing and two-dimensional electrophoresis: application to the RB1 gene. Hum Mol Genet. 1996 Jun;5(6):755–761. doi: 10.1093/hmg/5.6.755. [DOI] [PubMed] [Google Scholar]
- Weng A., Engler P., Storb U. The bulk chromatin structure of a murine transgene does not vary with its transcriptional or DNA methylation status. Mol Cell Biol. 1995 Jan;15(1):572–579. doi: 10.1128/mcb.15.1.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacksenhaus E., Bremner R., Phillips R. A., Gallie B. L. A bipartite nuclear localization signal in the retinoblastoma gene product and its importance for biological activity. Mol Cell Biol. 1993 Aug;13(8):4588–4599. doi: 10.1128/mcb.13.8.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]