Abstract
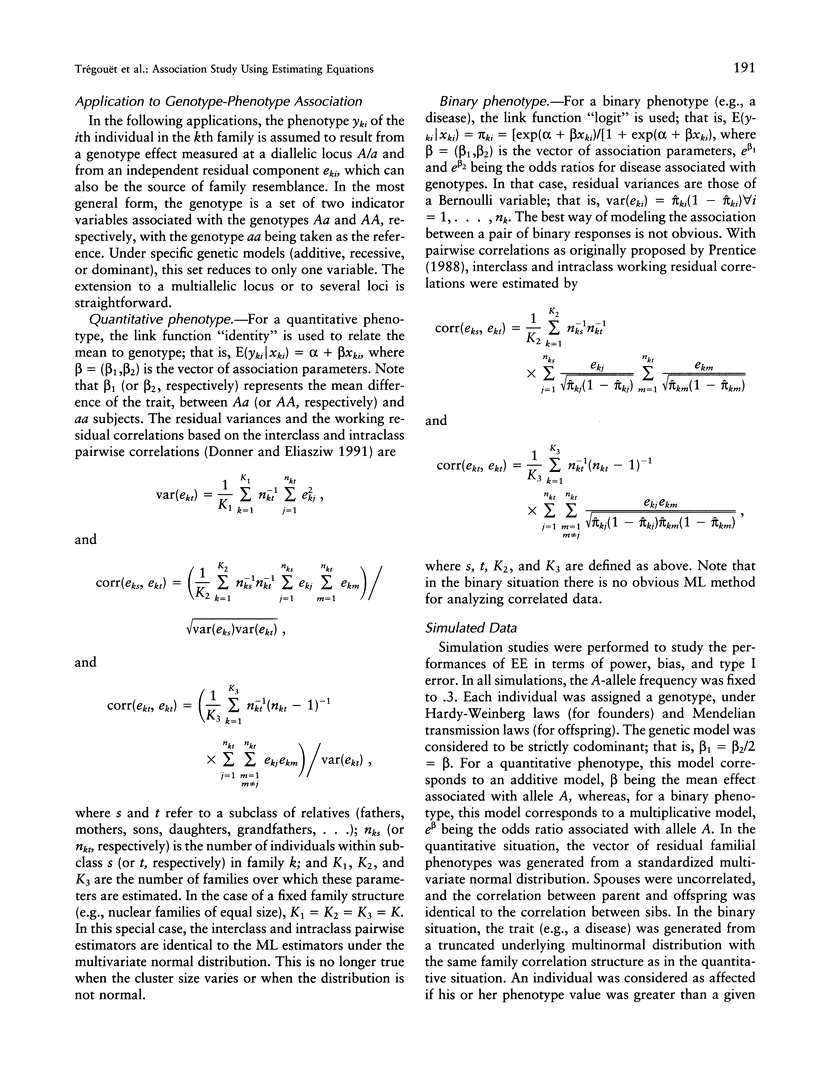
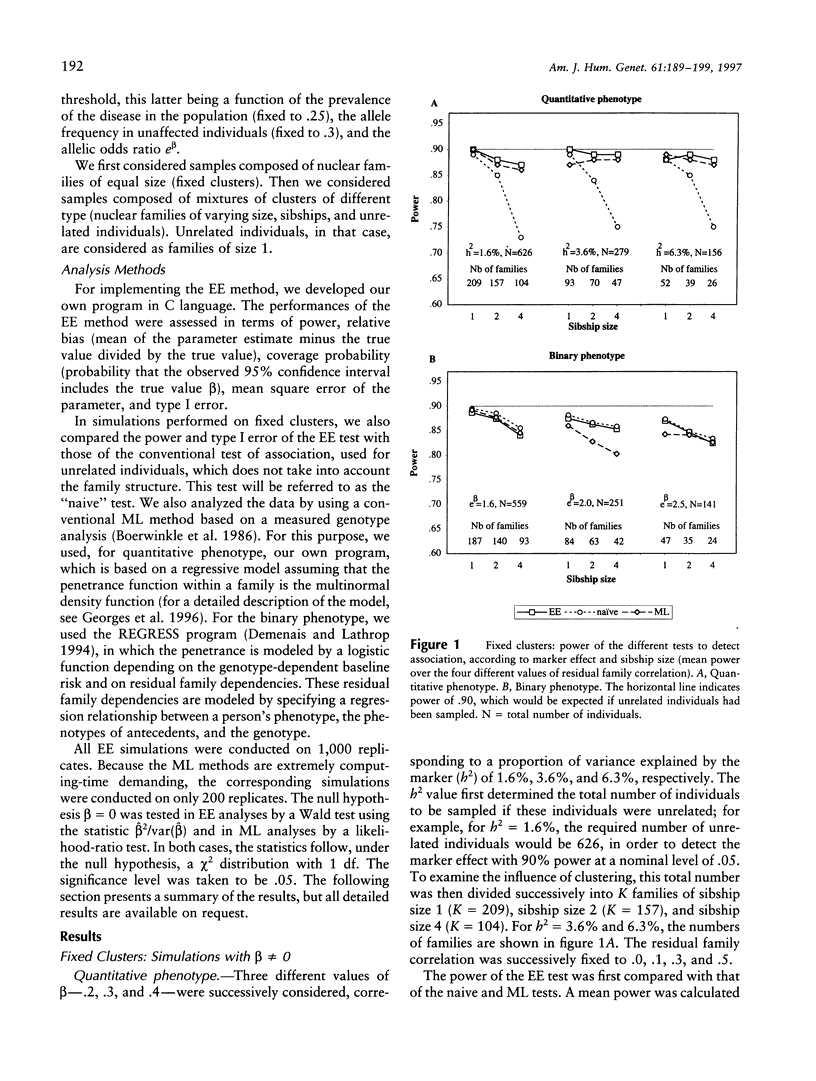
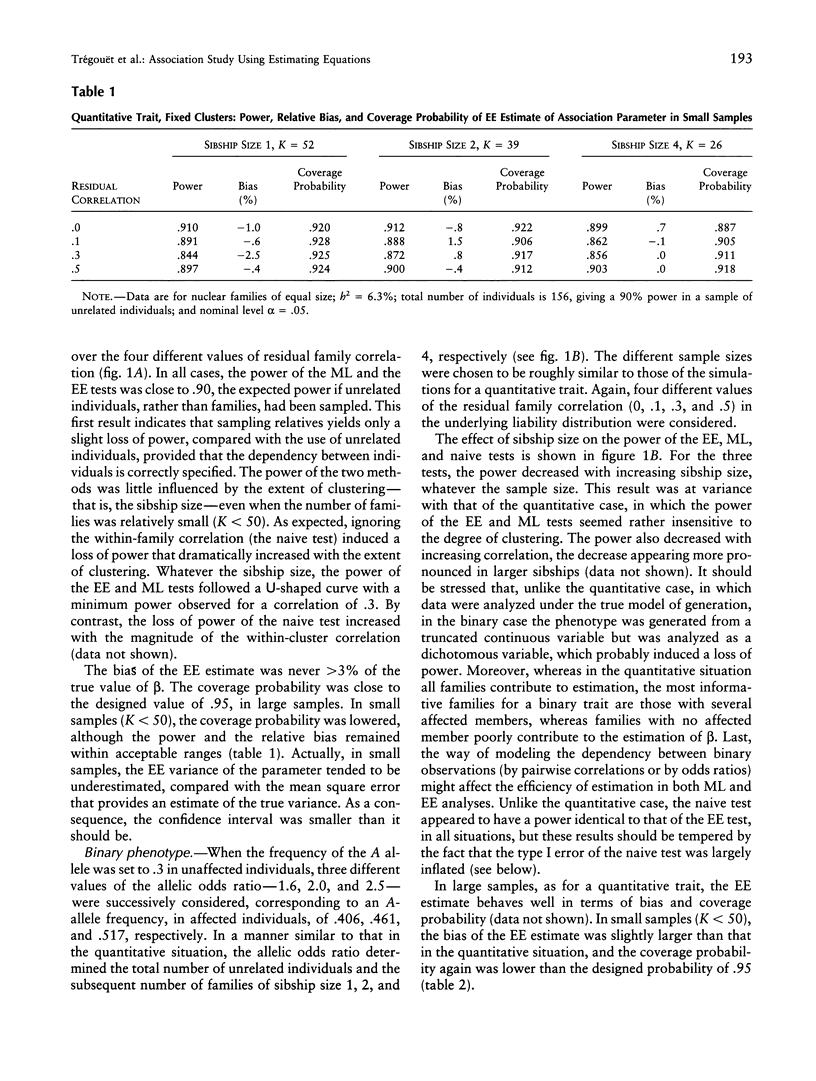
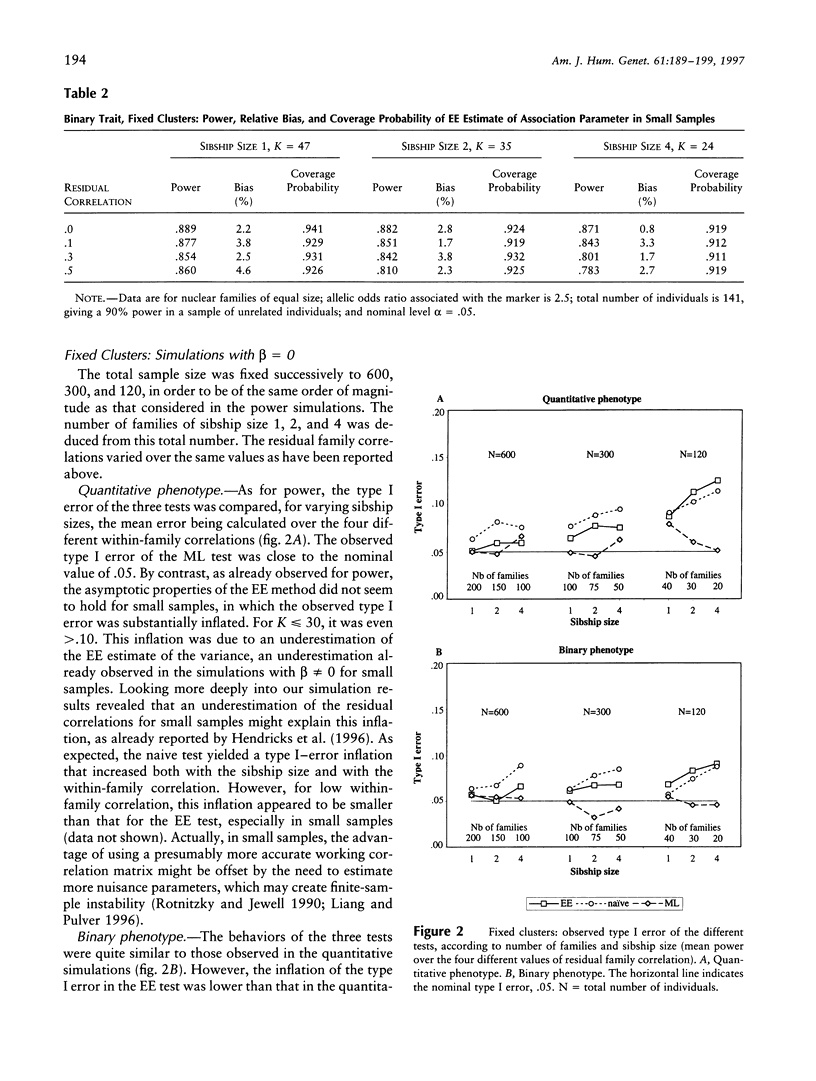
Association studies are one of the major strategies for identifying genetic factors underlying complex traits. In samples of related individuals, conventional statistical procedures are not valid for testing association, and maximum likelihood (ML) methods have to be used, but they are computationally demanding and are not necessarily robust to violations of their assumptions. Estimating equations (EE) offer an alternative to ML methods, for estimating association parameters in correlated data. We studied through simulations the behavior of EE in a large range of practical situations, including samples of nuclear families of varying sizes and mixtures of related and unrelated individuals. For a quantitative phenotype, the power of the EE test was comparable to that of a conventional ML test and close to the power expected in a sample of unrelated individuals. For a binary phenotype, the power of the EE test decreased with the degree of clustering, as did the power of the ML test. This result might be partly explained by a modeling of the correlations between responses that is less efficient than that in the quantitative case. In small samples (< 50 families), the variance of the EE association parameter tended to be underestimated, leading to an inflation of the type I error. The heterogeneity of cluster size induced a slight loss of efficiency of the EE estimator, by comparison with balanced samples. The major advantages of the EE technique are its computational simplicity and its great flexibility, easily allowing investigation of gene-gene and gene-environment interactions. It constitutes a powerful tool for testing genotype-phenotype association in related individuals.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abel L., Golmard J. L., Mallet A. An autologistic model for the genetic analysis of familial binary data. Am J Hum Genet. 1993 Oct;53(4):894–907. [PMC free article] [PubMed] [Google Scholar]
- Boerwinkle E., Chakraborty R., Sing C. F. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986 May;50(Pt 2):181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- Bonnardeaux A., Davies E., Jeunemaitre X., Féry I., Charru A., Clauser E., Tiret L., Cambien F., Corvol P., Soubrier F. Angiotensin II type 1 receptor gene polymorphisms in human essential hypertension. Hypertension. 1994 Jul;24(1):63–69. doi: 10.1161/01.hyp.24.1.63. [DOI] [PubMed] [Google Scholar]
- Bonney G. E. Compound regressive models for family data. Hum Hered. 1992;42(1):28–41. doi: 10.1159/000154044. [DOI] [PubMed] [Google Scholar]
- Bull S. B., Chapman N. H., Greenwood C. M., Darlington G. A. Evaluation of genetic and environmental effects using GEE and APM methods. Genet Epidemiol. 1995;12(6):729–734. doi: 10.1002/gepi.1370120633. [DOI] [PubMed] [Google Scholar]
- Demenais F. M. Regressive logistic models for familial diseases: a formulation assuming an underlying liability model. Am J Hum Genet. 1991 Oct;49(4):773–785. [PMC free article] [PubMed] [Google Scholar]
- Donner A., Eliasziw M. Methodology for inferences concerning familial correlations: a review. J Clin Epidemiol. 1991;44(4-5):449–455. doi: 10.1016/0895-4356(91)90084-m. [DOI] [PubMed] [Google Scholar]
- Garbers D. L. Guanylyl cyclase receptors and their endocrine, paracrine, and autocrine ligands. Cell. 1992 Oct 2;71(1):1–4. doi: 10.1016/0092-8674(92)90258-e. [DOI] [PubMed] [Google Scholar]
- Georges J. L., Régis-Bailly A., Salah D., Rakotovao R., Siest G., Visvikis S., Tiret L. Family study of lipoprotein lipase gene polymorphisms and plasma triglyceride levels. Genet Epidemiol. 1996;13(2):179–192. doi: 10.1002/(SICI)1098-2272(1996)13:2<179::AID-GEPI4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Grove J. S., Zhao L. P., Quiaoit F. Correlation analysis of twin data with repeated measures based on generalized estimating equations. Genet Epidemiol. 1993;10(6):539–544. doi: 10.1002/gepi.1370100635. [DOI] [PubMed] [Google Scholar]
- Hendricks S. A., Wassell J. T., Collins J. W., Sedlak S. L. Power determination for geographically clustered data using generalized estimating equations. Stat Med. 1996 Sep 15;15(17-18):1951–1960. doi: 10.1002/(sici)1097-0258(19960930)15:18<1951::aid-sim407>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Hsu L., Zhao L. P. Assessing familial aggregation of age at onset, by using estimating equations, with application to breast cancer. Am J Hum Genet. 1996 May;58(5):1057–1071. [PMC free article] [PubMed] [Google Scholar]
- Lander E. S. The new genomics: global views of biology. Science. 1996 Oct 25;274(5287):536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- Lee H., Stram D. O. Segregation analysis of continuous phenotypes by using higher sample moments. Am J Hum Genet. 1996 Jan;58(1):213–224. [PMC free article] [PubMed] [Google Scholar]
- Lee H., Stram D. O., Thomas D. C. A generalized estimating equations approach to fitting major gene models in segregation analysis of continuous phenotypes. Genet Epidemiol. 1993;10(1):61–74. doi: 10.1002/gepi.1370100107. [DOI] [PubMed] [Google Scholar]
- Liang K. Y., Beaty T. H. Measuring familial aggregation by using odds-ratio regression models. Genet Epidemiol. 1991;8(6):361–370. doi: 10.1002/gepi.1370080602. [DOI] [PubMed] [Google Scholar]
- Liang K. Y., Pulver A. E. Analysis of case-control/family sampling design. Genet Epidemiol. 1996;13(3):253–270. doi: 10.1002/(SICI)1098-2272(1996)13:3<253::AID-GEPI3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Lindpaintner K., Lee M., Larson M. G., Rao V. S., Pfeffer M. A., Ordovas J. M., Schaefer E. J., Wilson A. F., Wilson P. W., Vasan R. S. Absence of association or genetic linkage between the angiotensin-converting-enzyme gene and left ventricular mass. N Engl J Med. 1996 Apr 18;334(16):1023–1028. doi: 10.1056/NEJM199604183341604. [DOI] [PubMed] [Google Scholar]
- Lipsitz S. R., Fitzmaurice G. M. Estimating equations for measures of association between repeated binary responses. Biometrics. 1996 Sep;52(3):903–912. [PubMed] [Google Scholar]
- Mancl L. A., Leroux B. G. Efficiency of regression estimates for clustered data. Biometrics. 1996 Jun;52(2):500–511. [PubMed] [Google Scholar]
- Olson J. M. Robust estimation of gene frequency and association parameters. Biometrics. 1994 Sep;50(3):665–674. [PubMed] [Google Scholar]
- Olson J. M., Wijsman E. M. Linkage between quantitative trait and marker loci: methods using all relative pairs. Genet Epidemiol. 1993;10(2):87–102. doi: 10.1002/gepi.1370100202. [DOI] [PubMed] [Google Scholar]
- Park T. A comparison of the generalized estimating equation approach with the maximum likelihood approach for repeated measurements. Stat Med. 1993 Sep 30;12(18):1723–1732. doi: 10.1002/sim.4780121807. [DOI] [PubMed] [Google Scholar]
- Prentice R. L. Correlated binary regression with covariates specific to each binary observation. Biometrics. 1988 Dec;44(4):1033–1048. [PubMed] [Google Scholar]
- Rice T., Province M., Pérusse L., Bouchard C., Rao D. C. Cross-trait familial resemblance for body fat and blood pressure: familial correlations in the Québec Family Study. Am J Hum Genet. 1994 Nov;55(5):1019–1029. [PMC free article] [PubMed] [Google Scholar]
- Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996 Sep 13;273(5281):1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Stram D. O., Lee H., Thomas D. C. Use of generalized estimating equations in segregation analysis of continuous outcomes. Genet Epidemiol. 1993;10(6):575–579. doi: 10.1002/gepi.1370100641. [DOI] [PubMed] [Google Scholar]
- Tiret L., Rigat B., Visvikis S., Breda C., Corvol P., Cambien F., Soubrier F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet. 1992 Jul;51(1):197–205. [PMC free article] [PubMed] [Google Scholar]
- Whittemore A. S., Gong G. Segregation analysis of case-control data using generalized estimating equations. Biometrics. 1994 Dec;50(4):1073–1087. [PubMed] [Google Scholar]
- Zhao L. P., Grove J. S. Identifiability of segregation parameters using estimating equations. Hum Hered. 1995 Sep-Oct;45(5):286–300. doi: 10.1159/000154315. [DOI] [PubMed] [Google Scholar]
- Zhao L. P., Grove J., Quiaoit F. A method for assessing patterns of familial resemblance in complex human pedigrees, with an application to the nevus-count data in Utah kindreds. Am J Hum Genet. 1992 Jul;51(1):178–190. [PMC free article] [PubMed] [Google Scholar]


