Abstract
We recently reported the identification of recurrent gene fusions in the majority of prostate cancers involving the 5′ untranslated region of the androgenregulated gene TMPRSS2 and the ETS family members ERG, ETV1, and ETV4. Here we report the noninvasive detection of these gene fusions in the urine of patients with clinically localized prostate cancer. By quantitative polymerase chain reaction, we assessed the expression of ERG and TMPRSS2:ERG transcripts in urine samples obtained after prostatic massage from 19 patients (11 prebiopsy and 8 pre-radical prostatectomy) with prostate cancer. We observed a strong concordance between ERG overexpression and TMPRSS2:ERG expression, with 8 of 19 (42%) patients having detectable TMPRSS2:ERG transcripts in their urine. Importantly, by fluorescence in situ hybridization, we confirmed the presence or the absence of TMPRSS2:ERG gene fusions in matched prostate cancer tissue samples from three of three patients with fusion transcripts in their urine and from two of two patients without fusion transcripts in their urine. These results demonstrate that TMPRSS2:ERG gene fusions can be detected in the urine of patients with prostate cancer and support larger studies on prospective cohorts for noninvasive detection of prostate cancer.
Keywords: Gene fusions, prostate cancer, noninvasive detection, urine, quantitative PCR
Introduction
Chromosomal rearrangements play causal roles in numerous human malignancies and have been exploited diagnostically and therapeutically [1,2]. Using a novel bioinformatics strategy to nominate candidate oncogenes, we identified recurrent gene fusions involving the 5′ untranslated region of the androgen-regulated gene TMPRSS2 to members of the ETS gene family (ERG, ETV1, or ETV4) in the majority of prostate cancers [3,4]. Subsequently, multiple studies have confirmed the presence of TMPRSS2:ETS gene fusions, particularly TMRPSS2:ERG, in 40% to 80% of prostate cancers [5–8]. In addition to likely playing a central role in the pathogenesis of prostate cancers, these studies highlight the potential of TMPRSS2:ETS gene fusions to serve as a specific biomarker of prostate cancer.
In an effort to develop a noninvasive method to detect TMPRSS2:ERG gene rearrangements, we explored the possibility of identifying this fusion in urine samples obtained from patients with prostate cancer using quantitative polymerase chain reaction (qPCR). Here we show that RNA isolated from sedimented urine and subjected to qPCR revealed the presence of TMPRSS2:ERG fusions in 8 of 19 (42%) patients with prostate cancer. We validated the specificity of this assay by confirming the presence or the absence of TMPRSS2:ERG gene rearrangements in matched tissue samples from a subset of our cohort. The results demonstrate the feasibility of the noninvasive detection of TMRPSS2:ETS gene fusions from the urine of patients with prostate cancer.
Materials and Methods
Urine Collection, RNA Isolation, and Amplification
This study was approved by the Institutional Review Board (IRB) of the University of Michigan Medical School (Ann Arbor, MI). With informed consent of the patients, urine samples were obtained following a digital rectal exam before either needle biopsy or radical prostatectomy. Urine was voided into urine collection cups containing DNA/RNA preservative (Sierra Diagnostics LLC, Sonora, CA). For RNA isolation, a minimum of 30 ml of urine was centrifuged at 4000 rpm for 15 minutes at 4°C. RNAlater (Ambion, Inc., Austin, TX) was added to urine sediments and stored at -20°C until RNA isolation. Total RNA was isolated using an RNeasy Micro kit (Qiagen, Inc., Valencia, CA) according to the manufacturer's instructions. RNA integrity was verified using an Agilent 2100 Bioanalyzer. Total RNA was amplified using an Omni-Plex Whole Transcriptome Amplification (WTA) kit (Rubicon Genomics, Ann Arbor, MI) according to the manufacturer's instructions, essentially as previously described [9]. Twenty-five nanograms of total RNA was used for WTA library synthesis, and cDNA library was subjected to one round of WTA PCR amplification. Amplified cDNA was purified using a QIAquick PCR Purification kit (Qiagen, Inc.). For cell line proof-of-concept experiments, the indicated number of VCaP or LNCaP cells was spiked into 1 ml of urine, and samples were processed like voided urine.
qPCR
qPCR was used to detect ERG, ETV1, and TMPRSS2:ERG transcripts from WTA-amplified cDNA, essentially as described [4]. For each qPCR, 10 ng of WTA-amplified cDNA was used as template. 2x Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA) and 25 ng of both forward and reverse primers were used for ERG, ETV1, prostate-specific antigen (PSA), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) qPCR. 2x Taqman Universal PCR Master Mix, a final concentration of 900 nM forward and reverse primers, and 250 nM probe were used for Taqman TMPRSS2:ERGa. For the Taqman assay, samples with Ct (threshold cycle) values greater than 38 cycles were considered to show no amplification. Threshold levels were set at the exponential phase of qPCR using Sequence Detection Software version 1.2.2 (Applied Biosystems). The amount of each target gene relative to the housekeeping gene GAPDH for each sample was determined using the comparative threshold cycle (Ct) method (Applied Biosystems user bulletin 2; http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf). Samples with inadequate amplification of PSA (Ct > 22), indicating poor recovery of prostate cells in the urine, were excluded from further analysis. ERG (exons 5 and 6) and ETV1 (exons 6 and 7) [4], GAPDH [10], and PSA [11] primers were as described. All primers were synthesized by Integrated DNA Technologies (Coralville, IA). Taqman primers and probe (MGB-labeled, synthesized by Applied Biosystems) specific for TMPRSS2:ERGa are as follows:
TM-ERGa2_MGB-f: CGCGGCAGGAAGCCTTA TM-ERGa2_MGB-r: TCCGTAGGCACACTCAAACAAC TM-ERGa2_MGB-probe: 5′-MGB-CAGTTGTGAGTGAGGACC- NFQ-3′.
Fluorescence In Situ Hybridization (FISH)
We acquired matched biopsy tissues from the University of Michigan Prostate Cancer Specialized Program of Research Excellence (SPORE) Tissue Core and prostatectomy tissue sections from the radical prostatectomy series at the University of Michigan, which is part of the SPORE. All samples were collected with informed consent of the patients and prior IRB approval. Formalin-fixed paraffin-embedded tissue sections were used for interphase FISH, as described [3,4]. For metaphase FISH on VCaP and LNCaP cells, metaphase spreads were prepared using standard methods. For analysis of ERG gene rearrangement, we used a split-signal probe strategy, with two probes spanning the ERG locus (5′, digoxin dUTP-labeled BAC clone RP11-95I21; 3′, biotin 14-dCTP-labeled BAC clone RP11-476D17). All BAC clones were obtained from the Children's Hospital of Oakland Research Institute.
Results and Discussion
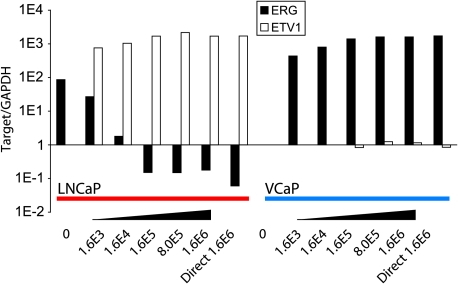
We sought to develop a method to detect the presence of TMPRSS2:ETS fusion transcripts in prostate cancer cells shed into the urine after a digital rectal exam. As proof of concept, we employed urine spiked with prostate cancer cell lines expressing high levels of ERG and TMPRSS2:ERG (VCaP) or high levels of ETV1 (LNCaP). As shown in Figure 1, we were able to detect ERG overexpression exclusively in VCaP at 1600 cells and ETV1 overexpression exclusively in LNCaP at 16000 cells by qPCR. By correlating the number of spiked VCaP and LNCaP cells to GAPDH and PSA Ct values, we observed that urine obtained from patients after a digital rectal exam contained cell numbers insufficient to reliably detect ERG or ETV1 overexpression (data not shown). Thus, we amplified total RNA collected from the urine of patients with prostate cancer using Omni-Plex WTA before qPCR analysis. We have previously validated WTA for RNA amplification before qPCR and/or DNA microarray analysis [9]. Using this strategy, we assessed two cohorts containing a total of 19 men with prostate cancer. After a digital rectal exam, urine was collected from 11 men before the performance of needle biopsy, which revealed the presence of prostate cancer. We also assessed a cohort of eight patients with prostate cancer from whom urine was collected after a digital rectal exam but before radical prostatectomy. Cohort characteristics are presented in Table 1.
Figure 1.
Detection of ERG and ETV1 transcripts in urine spiked with prostate cancer cell lines. The indicated number of LNCaP (red bar: high ETV1 expression) or VCaP (blue bar: high ERG and TMPRSS2:ERG expression) prostate cancer cells was spiked into 1 ml of urine. Approximately 1.6 million cells of each cell line were used without being spiked (Direct). Total RNA was isolated and reverse-transcribed to cDNA before qPCR analysis. The relative amount of ERG and ETV1 for each sample was normalized to the amount of GAPDH.
Table 1.
Noninvasive Detection of TMPRSS2:ERG Gene Fusions in the Urine of Men with Prostate Cancer.
| Sample ID | Type | Gleason Major | Gleason Minor | Gleason Score | PSA (ng/ml) | Age | ERG | Fusion (Ct) | FISH |
| LNCaP Cell line | NA | NA | NA | NA | NA | 0.01 | NQ | - | |
| 5778 | Bx | 3 | 3 | 6 | 11.7 | 52 | NQ | NQ | - |
| 5797 | RP | 3 | 4 | 7 | 5.3 | 52 | NQ | NQ | - |
| 5892 | RP | 4 | 3 | 7 | 8.9 | 57 | NQ | NQ | |
| 5909 | Bx | 3 | 3 | 6 | 7.8 | 56 | 0.04 | NQ | |
| 5918 | Bx | 3 | 3 | 6 | 3 | 56 | 0.06 | NQ | |
| 5915 | Bx | 3 | 4 | 7 | 6.9 | 71 | 0.20 | NQ | |
| 5798 | RP | 3 | 3 | 6 | 2.7 | 47 | 0.20 | NQ | |
| 5859 | RP | 3 | 3 | 6 | 8.7 | 63 | 0.27 | NQ | |
| 5893 | RP | 3 | 3 | 6 | 0.22 | 59 | 0.38 | 33.71 | |
| 5880 | RP | 3 | 3 | 6 | 2.96 | 67 | 0.40 | NQ | |
| 5796 | Bx | 4 | 5 | 9 | 19.3 | 82 | 0.98 | NQ | |
| 5780 | Bx | 3 | 3 | 6 | 5.9 | 79 | 1.00 | NQ | |
| 5794 | Bx | 3 | 3 | 6 | 3.8 | 56 | 1.09 | 38.96 | |
| 5864 | RP | 3 | 4 | 7 | 5.5 | 49 | 18.06 | 32.65 | |
| 5776 | Bx | 3 | 3 | 6 | 2.8 | 54 | 22.01 | 30.66 | + |
| 5775 | Bx | 3 | 3 | 6 | 5.99 | 62 | 30.91 | 32.87 | |
| 5815 | RP | 3 | 4 | 7 | 5.4 | 59 | 206.50 | 31.78 | + |
| 5790 | Bx | 3 | 4 | 7 | 5.5 | 51 | 328.56 | 31.48 | + |
| 5912 | Bx | 3 | 4 | 7 | 15.5 | 67 | 797.86 | 34.13 | |
| VCaP Cell line | NA | NA | NA | NA | NA | 226633.25 | 21.66 | + |
Each urine specimen was obtained from a unique patient assigned an ID, and urine samples spiked with 1.6 million VCaP or LNCaP cells were also assessed. The source of the sample, prebiopsy (Bx) or pre-radical prostatectomy (RP), is indicated. For all patients, major Gleason, minor Gleason, Gleason sum score, prebiopsy or preprostatectomy PSA (ng/ml), and age are reported. qPCR was used to measure the amount of ERG relative to PSA for each specimen. Samples were also assessed for the expression of TMPRSS2:ERGa using a specific Taqman assay, with positive samples indicated by the threshold cycle (Ct) of amplification. Matched prostate cancer tissue samples for five samples were assessed by FISH for TMPRSS2:ERG fusion using a split-probe assay for ERG rearrangement. Samples negative or positive for TMPRSS2:ERG rearrangements are indicated by (-) or (+), respectively. NQ, no quantifiable amplification of ERG; ND, no detectable amplification of ERG or TMPRSS2:ERG for the respective assays.
For each patient, we determined the expression of ERG relative to PSA, in addition to determining whether the sample expressed TMPRSS2:ERG fusion transcripts. To confirm the specificity of our TMPRSS2:ERG Taqman primer/probe assay, we assayed urine samples spiked with 1.6 million LNCaP or VCaP cells. We detected TMPRSS2:ERG fusion transcripts exclusively in VCaP cells, which we have previously shown to markedly overexpress ERG and to harbor TMPRSS2:ERG rearrangement [4]. By this same assay, 8 of 19 (42%) urine samples expressed TMPRSS2:ERG, including the seven samples with the highest expression of ERG (Table 1). These results are consistent with previous studies demonstrating an overall frequency of 40% to 80% for TMPRSS2:ERG fusions in prostate cancer tissue samples and demonstrating that ∼95% of samples with ERG overexpression harbor TMPRSS2:ERG gene fusions [4–8]. We did not detect ETV1 overexpression in any sample.
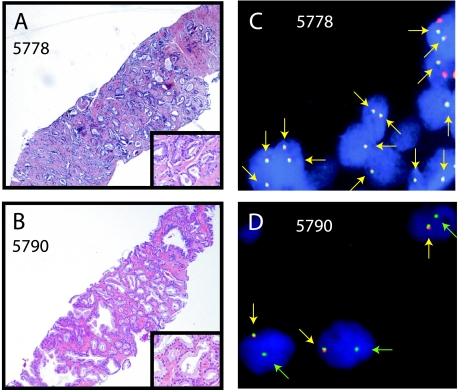
As a confirmation of the specificity of our qPCR assay, we used FISH on matched tissue samples to determine the presence or the absence of the TMPRSS2:ERG gene rearrangement in the patient's prostate cancer.Weused a splitprobe FISH assay, with probes located 5′ and 3′ to the ERG, where a TMPRSS2:ERG gene rearrangement is indicated by splitting of one pair of probes or by loss of the 5′ ERG probe, which is consistent with an intrachromosomal deletion between TMPRSS2 and ERG on chromosome 21q [4,5,8]. We expected that prostate cancer tissues from patients with high levels of ERG and TMPRSS2:ERG transcripts in their urine should be positive by FISH, whereas prostate cancer from patients with low levels of ERG and no detectable TMPRSS2:ERG transcripts in their urine should be negative by FISH. Thus, we assessed matched prostate tissue samples from three patients with detectable TMPRSS2:ERG in their urine and from two patients without detectable TMPRSS2:ERG in their urine. As expected, tissues from the three patients with high levels of ERG and detectable levels of TMRPSS2:ERG in their urine were positive for ERG rearrangement by FISH, whereas the two samples without TMPRSS2:ERG in their urine were negative for ERG rearrangement by FISH (Table 1). Hematoxylin and eosin-stained tissue sections and corresponding negative FISH assay from sample 5778, and a positive FISH assay from sample 5790 with deletion of the 5′ ERG probe are shown in Figure 2, A–D.
Figure 2.
Confirmation of the presence or the absence of TMPRSS2:ERG detection in the urine using FISH on matched tissue sections. Matched prostate cancer tissue samples for five samples were assessed by FISH for TMPRSS2:ERG fusion using a split-probe assay for ERG rearrangement (Table 1). Hematoxylin and eosin staining (A and B) and representative FISH images (C and D) for samples 5778 and 5790 are shown. A negative FISH assay (C) for sample 5778 is indicated by two pairs of colocalized red and green signals (yellow arrows) per cell, whereas a positive FISH assay is indicated by one pair of split red and green signals (not shown) or exclusive loss of the 5′ ERG probe (red signal) resulting in one pair of colocalized signals (yellow arrows) and one green signal (green arrows) per cell (D), as shown for sample 5790.
In summary, we have described the noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of patients with prostate cancer. We and others have recently described the presence of TMPRSS2:ETS gene fusions in the majority of prostate cancers and the utility of these gene fusions as a specific tissue biomarker of prostate cancer [3–8]. One limitation of the TMPRSS2:ERG Taqman assay we used for this study is that it only detects the TMPRSS2:ERGa isoform, which is expressed in approximately 85% to 95% of fusion-positive prostate cancers [4,7]. Thus, additional assays will be needed to detect alternative isoforms expressed in the remaining 10% to 20% of positive cases. Isoform-specific assays may be particularly relevant, as particular isoforms have been associated with aggressive disease [7]. The presence of prostate cancer cells in the sedimented urine of prostate cancer suggests that other approaches to detect TMPRSS2:ETS gene rearrangements, such as urine-based FISH similar to the UroVysion system for detecting bladder cancer [12], may also be feasible. A FISH-based assay would also be able to identify TMPRSS2:ERG+ cases with intrachromosomal deletion between TMPRSS2 and ERG, which has also been associated with aggressive disease in some cohorts [5,8]. In conclusion, the results reported herein support large-scale studies in prospective cohorts to determine the specificity and the sensitivity of urine-based assays for the detection of prostate cancer.
Acknowledgements
The authors thank Bo Han, Anjana Menon, and Alex Bond for technical assistance.
Footnotes
This work was supported, in part, by the National Institutes of Health Early Detection Research Network (UO1 CA111275-01 to A.M.C. and J.T.W.), the Prostate SPORE (P50CA69568 to K.JA.M.C. and R.B.S.), the Prostate Cancer Foundation (to A.M.C.), and the Department of Defense (PC040517 to R.M.). S.A.T. is supported by a Rackham Predoctoral Fellowship and is a fellow of the Medical Scientist Training Program. A.M.C. is supported by a Clinical Translational Research Award from the Burroughs Welcome Foundation.
Co-first authors.
References
- 1.Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 2.Rowley JD. Chromosome translocations: dangerous liaisons revisited. Nat Rev Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 3.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 4.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 5.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 6.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45:717–719. doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 8.Yoshimoto M, Joshua AM, Chilton-Macneill S, Bayani J, Selvarajah S, Evans AJ, Zielenska M, Squire JA. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia. 2006;8:465–469. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlins SA, Mehra R, Rhodes DR, Shah RB, Rubin MA, Bruening E, Makarov V, Chinnaiyan AM. Whole transcriptome amplification for gene expression profiling and development of molecular archives. Neoplasia. 2006;8:153–162. doi: 10.1593/neo.05754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Specht K, Richter T, Muller U, Walch A, Werner M, Hofler H. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158:419–429. doi: 10.1016/S0002-9440(10)63985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halling KC. Vysis UroVysion for the detection of urothelial carcinoma. Expert Rev Mol Diagn. 2003;3:507–519. doi: 10.1586/14737159.3.4.507. [DOI] [PubMed] [Google Scholar]