Abstract
TMPRSS2-ETS gene fusions have been found recurrently in prostate carcinomas, but not in the presumed precursor lesion, high-grade prostatic intraepithelial neoplasia (HGPIN). However, HGPIN lesions may share chromosomal changes with prostate cancer. To determine the relative order of genetic events in prostate carcinogenesis, we have analyzed 34 prostate carcinomas, 19 paired HGPIN lesions, 14 benign prostate hyperplasias, and 11 morphologically normal prostatic tissues for TMPRSS2-ERG and TMPRSS2-ETV1 rearrangements and genomic imbalances. TMPRSS2 exon 1 was fused in-frame with ERG exon 4 in 17 of 34 (50%) prostate carcinomas and in 4 of 19 (21%) HGPIN lesions, but in none of controls. The findings were further validated by sequencing analysis and by the real-time polymerase chain reaction quantification of TMPRSS2-ERG fusion transcript and the ERG exons 5/6:exons 1/2 expression ratio. Chromosome copy number changes were detected by comparative genomic hybridization in 42% of clinically confined carcinomas and in none of the 16 HGPIN lesions analyzed. We demonstrate for the first time that the TMPRSS2-ERG fusion gene can be detected in a proportion of HGPIN lesions and that this molecular rearrangement is an early event that may precede chromosome-level alterations in prostate carcinogenesis.
Keywords: TMPRSS2-ETS fusion oncogenes, prostate cancer, high-grade prostatic intraepithelial neoplasia, chromosomal changes, ERG
Introduction
A central aim in cancer research is to identify genes that play a causal role in cancer development. Many such genes have been identified through analyses of recurrent chromosomal rearrangements that are characteristic of leukemias, lymphomas, and sarcomas, typically resulting in the formation of oncogenic fusion genes [1]. This type of specific genetic change has only rarely been detected in common solid cancers [2], although that can be related to the smaller number of cases analyzed [3,4]. Recently, taking advantage of a bioinformatic approach termed Cancer Outlier Profile Analysis, the fusion genes TMPRSS2-ERG and TMPRSS2-ETV1 have been detected in a high proportion of prostate carcinomas selected for demonstrating an overexpression of the erythroblast transformation specific (ETS) transcription factor ERG or ETV1 [5]. Later, a rare third fusion gene, involving the TMPRSS2 locus and another ETS family gene ETV4, was identified [6].
The fusion partner common to the three rearrangements is TMPRSS2, an androgen-regulated member of the type II transmembrane serine protease family that maps to 21q22.3. TMPRSS2 protein is preferentially expressed in normal prostate tissues and is overexpressed in the neoplastic prostatic epithelium [7–10]. Its expression seems to be regulated by androgen-responsive elements (AREs) in a promoter [10,11], and it has been shown that androgen stimulation can induce the overexpression of ERG in a TMPRSS2-ERG-positive cell line [5]. These results suggest that deregulation of ETS transcription factor protein activity through AREs mapped 5′ of TMPRSS2 may underlie prostate cancer development, affecting biologic processes such as cell proliferation, differentiation, development, transformation, and apoptosis [5,12].
Although TMPRSS2-ETS gene fusions seem to be recurrent in prostate carcinomas, this genetic abnormality has not been reported in the presumed precursor lesion, high-grade prostatic intraepithelial neoplasia (HGPIN) [5]. However, fluorescent in situ hybridization (FISH) and comparative genomic hybridization (CGH) data have shown that HGPIN lesions may share genetic features with prostate cancer (e.g., 8p deletion) [13,14]. Therefore, the time of occurrence and the relative order of events in ETS gene fusions and chromosome imbalances are not known in prostate carcinogenesis. To address this issue, we have analyzed 34 samples of clinically localized prostate adenocarcinomas (PCa) and 19 paired HGPIN lesions for chromosome copy number changes and TMPRSS2-ERG and TMPRSS2-ETV1 rearrangements.
Materials and Methods
Patient Data
Primary tumors from 34 patients with clinically localized PCa [stage II (T1cN0M0 or T2N0M0), according to the TNM staging system] who were consecutively diagnosed and primarily treated with radical prostatectomy at the Portuguese Oncology Institute (Porto, Portugal) were prospectively collected. In 19 radical prostatectomy specimens with PCa, HGPIN lesions were identified and collected for further analysis. For control purposes, non-neoplastic prostate tissue samples were obtained from 14 randomly selected patients with benign prostate hyperplasia (BPH) who underwent transurethral resection of the prostate and from the peripheral zone of 11 prostates that did not harbor prostate cancer, which were collected from cystoprostatectomy (NPT) specimens of bladder cancer patients.
Sample Collection, RNA Extraction, and cDNA Synthesis
All tissue specimens were frozen immediately after surgery and stored at -80°C for further analysis. Five-micron-thick sections were cut and stained for the identification of areas of PCa (i.e., index or dominant tumor), HGPIN, BPH, and morphologically normal tissues. Then, the tissue block was trimmed to maximize the yield of target cells (> 70% of target cells). Subsequently, an average of fifty 12-µm-thick sections were cut, and every fifth section was stained to ensure a uniform percentage of target cells and to exclude contamination from neoplastic cells in normal and BPH tissue samples. Total cellular RNA was extracted from 250 mg of (normal and tumor) tissues using the FastRNA Kit Green (Qbiogene, Carlsbad, CA) for 90 seconds, with a speed rating of 6.0 in a FastPrep FP120 Instrument (Qbiogene). For cDNA synthesis, 1 to 5 µg of RNA was subjected to reverse transcription with random hexamers using the Superscript III First-Strand Synthesis System for reverse transcriptase-polymerase chain reaction (RT-PCR) (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. Final cDNA was diluted with 30 µl of H2O.
RT-PCR Analysis
RT-PCR for the detection of TMPRSS2-ERG and TMPRSS2-ETV1 chimeric transcripts was previously described [5]. In brief, PCR was performed in a 50-µl reaction containing 2 µl of synthesized cDNA, 5 µl of 10 x GeneAmp PCR Buffer II (100 mM Tris-HCl, pH 8.3, 500 mM KCl) (Applied Biosystems, Foster City, CA), 5 µl of 25 mM MgCl2, 0,4 µl of dNTP mix (25 mM of each dNTP) (Applied Biosystems), 0.4 µM of each primer (Metabion, Martinsried, Deutschland), and 1 U of AmpliTaq Gold DNA Polymerase (Applied Biosystems). Reaction tubes were kept on ice at all times to prevent nonspecific amplification. Reaction tubes were incubated for 10 minutes at 95°C, followed by 35 cycles of 1 minute at 95°C, 1 minute at 63°C, and 1 minute at 72°C, followed by a final elongation of 10 minutes at 72°C on a GeneAmp PCR System 9700 (Applied Biosystems). Amplified products were analyzed on a 2% agarose gel (SeaKem LE Agarose, Rockland, MA), and the results were visualized with an image analyzer ImageMaster VDS (Amersham Biosciences, Little Chalfont, UK).
Sequence Analysis
Sequence analysis was directly performed on amplified RT-PCR products with the use of BigDye Terminator Cycle Sequencing Chemistry (Applied Biosystems) on an automated sequencer ABI Prism 310 Genetic Analyzer (Applied Biosystems), according to the manufacturer's instructions.
Real-Time PCR Analysis
Primers and probes for TMPRSS2-ERG, TMPRSS2, ERG, and ETV1 were designed with Primer Express 2.0 (Applied Biosystems) and purchased from Metabion (Table 1). Primers and probes for the b-glucuronidase (GUSB) gene, used as endogenous control, were purchased as a predeveloped assay reagent from Applied Biosystems. To determine the relative expression levels of the target gene in each sample, the relative amount of the target gene was calibrated to the relative amount of the internal reference gene and was expressed in terms of target/reference ratios that were then multiplied by 100 for easier tabulation (target gene/GUSB x 100). PCR was performed in a 25-µl reaction containing 5 µl of synthesized cDNA, 12.5 µl of TaqMan universal PCR master mix, 0.3 µM of each primer, and 0.2 µM of each probe. PCR was performed in separate wells for each primer/probe set, and each sample was run in triplicate. PCR parameters were as follows: 50°C for 2 minutes, 95°C for 10 minutes, followed by 50 cycles at 95°C for 15 seconds and 60°C for 1 minute. Each plate included multiple nontemplate controls and serial dilutions of a positive control for constructing the standard curve.
Table 1.
Oligonucleotide Primers and Probes (5′FAM and 3′TAMRA) Used in This Study.
| Gene | Exon | Position | Primer/Probe* | Sequence 5′-3′ |
| ERG | 1 | 3–22 | ERG1-S | CCCGAGGGACATGAGAGAAG |
| ERG | 2 | 50–69 | ERG2-AS | TTTCCTCGGGTCTCCAAAGA |
| ERG | 1–2 | 26–48 | ERG12-PR | AGCGGCGCTCAGGTTATTCCAGG |
| ERG | 5 | 564–583 | ERG5-S | CACGAACGAGCGCAGAGTTA |
| ERG | 6 | 611–630 | ERG6-AS | CTGCCGCACATGGTCTGTAC |
| ERG | 5–6 | 585–609 | ERG56-PR | CGTGCCAGCAGATCCTACGCTATGG |
| TMPRSS2 | 1 | -4–17 | TMPRSS2/ERG-S | TAGGCGCGAGCTAAGCAGGAG |
| ERG | 4 | 252–276 | TMPRSS2/ERG-AS | GTAGGCACACTCAAACAACGACTGG |
| TMPRSS2-ERG | - | - | TMPRSS2/ERG-PR | AGCGCGGCAGGAAGCCTTATCAGTT |
The GenBank accession numbers for TMPRSS2 and ERG are NM_005656.2 and NM_004449.3, respectively.
S = sense; AS = antisense; PR = probe.
CGH Analysis
CGH analysis followed the procedure of Kallioniemi et al. [15], with modifications, as previously described [16]. Briefly, test and reference DNA were extracted using standard methods and labeled in nick translation reactions using Spectrum Green and Spectrum Red conjugated nucleotides (Vysis, Downers Grove, IL), after which probe lengths between 300 and 2000 bp were obtained. Labeled sample and reference DNA (1 µg each) were mixed with 30 µg of unlabeled Cot1 DNA (Life Technologies, Rockville, MD), ethanol-precipitated, dried, and dissolved in hybridization buffer (Vysis). Probe mixture was denatured and hybridized to commercially available normal metaphase slides (Vysis) for 2 to 3 days at 37°C in a moist chamber. After washing off excess probe, samples were counterstained with DAPI in an antifade solution (Vector Laboratories, Burlingame, CA). Image analysis was performed with CytoVision System version 3.0 (Applied Imaging, Santa Clara, CA). Data from 10 cells were combined to generate average ratio profiles with 99% confidence intervals (CIs) for each sample. A standard reference interval, generated with data from 10 normal versus normal hybridizations (totaling 110 cells), was automatically scaled onto each sample, and aberrations were scored whenever the case profile and the standard reference profile at 99% CI did not overlap [17]. The description of CGH copy number changes followed the guidelines suggested in ISCN [18].
Results
Frequency of TMPRSS2-ERG and TMPRSS2-ETV1 Fusion Transcripts
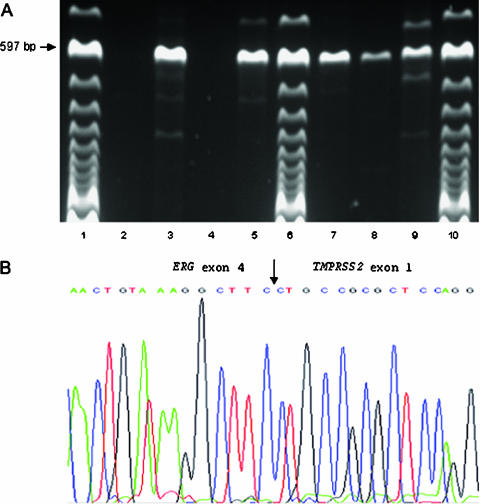
To estimate the frequency of TMPRSS2-ERG and TMPRSS2-ETV1 chimeric transcripts, we have screened a consecutive series of 34 patients with clinically localized PCa and 19 paired HGPIN. Type A TMPRSS2-ERG transcript could be detected in 17 of 34 (50%) prostate carcinomas and in 4 of 19 (21%) HGPIN lesions, but in none of controls (Figure 1A). When we consider all patient samples regardless of lesion type (i.e., PCa or HGPIN), this frequency rises to 56% (19 of 34) because, in two negative PCa cases, the corresponding HGPIN was positive (Table 2). No type B TMPRSS2-ERG or TMPRSS2-ETV1 fusion transcript was detected in any of the samples analyzed. The sequencing of amplification products, followed by BLAST search, confirmed that TMPRSS2 exon 1 was fused in-frame with ERG2 exon 4 (Figure 1B).
Figure 1.
Detection and analysis of type A TMPRSS2-ERG fusion transcript in prostate carcinoma and HGPIN samples. (A) RT-PCR analysis with a sense primer located in TMPRSS2 exon 1 and an antisense primer located in ERG exon 6 in PCa (PCa 55, PCa 114, PCa 134, and PCa 152 in lanes 2–5, respectively) and HGPIN (HGPIN 60, HGPIN 83, and HGPIN 42 in lanes 7–9, respectively) samples. The expected size of the type A TMPRSS2-ERG fusion transcript is indicated. Lanes 1, 6, and 10 = 100-bp molecular marker. (B) Partial sequence of the junction of type A TMPRSS2-ERG chimeric mRNA showing the nucleotide sequence of the fusion transcript. The arrow shows in-frame fusion between TMPRSS2 exon 1 and ERG exon 4.
Table 2.
Qualitative RT-PCR (Column 2), Quantitative RT-PCR (Columns 3 and 4), and CGH (Column 5) Findings of a Consecutive Series of PCa, Paired HGPIN, BPH, and NPT Samples.
| Sample ID | TMPRSS2-ERG RT-PCR | TMPRSS2-ERG Real-Time PCR | ERG Exons 5/6:Exons 1/2 Real-Time PCR | CGH Findings (Standard Reference Interval [SRI] 99% CI) |
| PCa 32 | Positive, type A | 19.42 | 4.77 | Rev ish dim(8p12pter) |
| HGPIN 32 | - | 0.00 | 0.44 | No changes |
| PCa 40 | Positive, type A | 530.63 | 52.45 | No changes |
| HGPIN 40 | - | 0.00 | 1.39 | No changes |
| PCa 42 | - | ND | 1.25 | No changes |
| HGPIN 42 | Positive, type A | 1.02 | 0.79 | No changes |
| PCa 45 | Positive, type A | 8.01 | 2.87 | Rev ish dim(8p12p22) |
| HGPIN 45 | - | ND | 0.37 | ND |
| PCa 46 | - | ND | 1.97 | ND |
| HGPIN 46 | - | ND | 0.41 | No changes |
| PCa 55 | - | ND | 2.29 | Rev ish enh(3q23q26,7p13p21,7q21q32,8q21q24), dim(16q22qter) |
| HGPIN 55 | - | 0.00 | 4.23 | No changes |
| PCa 56 | - | ND | 2.08 | No changes |
| PCa 58 | - | ND | 2.38 | Rev ish enh(8q21q24), dim(8p22) |
| HGPIN 58 | Positive, type A | 1.73 | 17.47 | No changes |
| PCa 60 | Positive, type A | 37.56 | 4.69 | No changes |
| HGPIN 60 | Positive, type A | 17.19 | 2.48 | No changes |
| PCa 67 | Positive, type A | 120.91 | 111.61 | Rev ish dim(8p21pter) |
| PCa 72 | - | ND | 0.44 | Rev ish enh(5p14pter,5q11q23,5q32q33) |
| PCa 76 | - | ND | 1.72 | Rev ish enh(8q), dim(2q23q24,8p12p23,10p11p12,10q22q25) |
| PCa 78 | - | ND | 1.73 | ND |
| HGPIN 78 | - | ND | 1.85 | No changes |
| PCa 81 | Positive, type A | 112.83 | 10.41 | No changes |
| PCa 83 | Positive, type A | 24.79 | 13.84 | No changes |
| HGPIN 83 | Positive, type A | 1.87 | 1.01 | ND |
| PCa 84 | - | 0.00 | 1.34 | No changes |
| HGPIN 84 | - | ND | 1.08 | No changes |
| PCa 87 | - | ND | 1.35 | No changes |
| HGPIN 87 | - | ND | 0.76 | No changes |
| PCa 89 | Positive, type A | 149.02 | 7.47 | No changes |
| PCa 101 | Positive, type A | 19.43 | 6.44 | Rev ish enh(18p11), dim(8p22pter,13q14q22) |
| PCa 114 | Positive, type A | 493.30 | 47.07 | Rev ish dim(16q22qter) |
| PCa 115 | - | ND | 2.17 | No changes |
| HGPIN 115 | - | 0.00 | 2.98 | No changes |
| PCa 131 | - | ND | 1.45 | No changes |
| PCa 134 | - | 0.00 | 1.18 | No changes |
| PCa 138 | Positive, type A | 13.58 | 31.49 | ND |
| HGPIN 138 | - | 0.00 | 0.29 | No changes |
| PCa 139 | Positive, type A | 62.40 | 59.25 | Rev ish dim(8p22pter,17p13) |
| PCa 140 | Positive, type A | 11.62 | 5.84 | No changes |
| PCa 145 | Positive, type A | 1.55 | 4.70 | No changes |
| HGPIN 145 | - | 0.00 | 1.68 | No changes |
| PCa 147 | - | ND | 2.24 | No changes |
| HGPIN 147 | - | ND | 0.57 | No changes |
| PCa 150 | - | ND | 0.53 | No changes |
| HGPIN 150 | - | ND | 1.03 | No changes |
| PCa 151 | - | 0.00 | 0.29 | No changes |
| PCa 152 | Positive, type A | 206.05 | 9.62 | No changes |
| PCa 156 | - | ND | 1.69 | Rev ish dim(6q15q21,8p21p23,13q21q31) |
| HGPIN 156 | - | 0.00 | 0.82 | ND |
| PCa 164 | Positive, type A | 57.74 | 6.34 | Rev ish dim(8p12p22,10q22qter,13q14q21,16q23q24) |
| HGPIN 164 | - | 0.00 | 0.42 | ND |
| PCa 172 | Positive, type A | 155.73 | 6.94 | Rev ish dim(8p12p22,17p12pter) |
| NPT 1 | - | ND | 0.84 | ND |
| NPT 3 | - | 0.00 | 2.08 | ND |
| NPT 5 | - | ND | 0.44 | ND |
| NPT 6 | - | ND | 0.45 | ND |
| NPT 7 | - | 0.00 | 1.96 | ND |
| NPT 8 | - | ND | 0.23 | ND |
| NPT 10 | - | ND | 0.07 | ND |
| NPT 11 | - | ND | 0.47 | ND |
| NPT 12 | - | 0.00 | 0.19 | ND |
| NPT 13 | - | 0.00 | 0.37 | ND |
| NPT 14 | - | 0.00 | 0.10 | ND |
| BPH 7 | - | ND | 0.70 | No changes |
| BPH 17 | - | ND | 0.32 | No changes |
| BPH 36 | - | ND | 0.12 | No changes |
| BPH 55 | - | ND | 0.38 | No changes |
| BPH 68 | - | ND | 0.06 | No changes |
| BPH 71 | - | ND | 0.08 | No changes |
| BPH 76 | - | ND | 0.35 | No changes |
| BPH 79 | - | 0.00 | 2.05 | No changes |
| BPH 84 | - | 0.00 | 24.68 | No changes |
| BPH 89 | - | 0.00 | 0.40 | No changes |
| BPH 91 | - | ND | 0.32 | No changes |
| BPH 92 | - | ND | 0.16 | No changes |
| BPH 96 | - | ND | 0.28 | No changes |
ND, not done.
TMPRSS2-ERG Fusion Transcript Quantification
To validate the findings regarding TMPRSS2-ERG, we designed a specific primer pair and probe for the type A TMPRSS2-ERG transcript. All RT-PCR-positive cases were quantified and, as additional negative controls, some TMPRSS2-ERG-negative cases were also analyzed. Only the previously identified RT-PCR-positive cases (PCa and HGPIN) showed amplification by real-time PCR, with TMPRSS2-ERG normalized values ranging from 1.55 to 530.63 in PCa and from 1.02 to 17.19 in HGPIN (Table 2). All TMPRSS2-ERG-negative cases analyzed showed no amplification by real-time PCR.
Relationship between TMPRSS2-ERG Fusion and ERG Overexpression
To evaluate the relationship between TMPRSS2-ERG detection and ERG expression in our series, we designed specific primer pairs and probes for ERG exons 1 and 2 and for ERG exons 5 and 6. Quantitative RT-PCR revealed that the ERG exons 5/6:exons 1/2 ratio was higher than 2.5 in all TMPRSS2-ERG-positive (but in none of negative) prostate carcinomas (Table 2). The relationship between the presence of TMPRSS2-ERG fusion and ERG overexpression was less constant in HGPIN lesions because only one of four positive cases for the TMPRSS2-ERG fusion showed an ERG exons 5/6:exons 1/2 ratio above 2.5. Regarding the control group, none of 11 NPT samples showed an ERG exons 5/6:exons 1/2 ratio above 2.5, but in one case of BPH (BPH 84), the ERG exons 5/6:exons 1/2 ratio was clearly above 2.5 (Table 2).
CGH Findings
Sixteen of 19 HGPIN lesions yielded enough DNA and were analyzed in the present study. Genomic data on 34 PCa and 14 BPH samples were previously published [14]. Chromosome copy number changes were detected in 42% of PCa samples (Table 2), with the most frequent alteration corresponding to 8p loss (10 of 13 samples with copy number changes). Of the 17 TMPRSS2-ERG-positive carcinomas, eight were shown to harbor genomic imbalances. All HGPIN lesions presented a balanced chromosome constitution. No statistically significant correlation could be established between specific genetic alterations and the presence of the TMPRSS2-ERG fusion transcript, but all three PCa with 8q gain did not harbor the fusion gene.
Discussion
We have confirmed that a high proportion of prostate carcinomas presents the TMPRSS2-ERG rearrangement. In our consecutive series of clinically localized PCa, this genetic anomaly was detected in 50% of cases, making it the most frequent fusion gene in human carcinomas. This finding was confirmed by quantitative real-time RT-PCR targeting the fusion gene. Furthermore, the ERG exons 5/6:exons 1/2 ratio was higher than 2.5 in all TMPRSS2-ERG-positive (but in none of negative) prostate carcinomas. Our results are in agreement with Tomlins et al. [5] who, using FISH analysis, reported the presence of ERG rearrangement in 55% (16 of 29) of a series of 29 prostate carcinomas selected independently of any knowledge of ERG or ETV1 expression [5]. However, Soller et al. [19] reported a TMPRSS2-ERG fusion frequency of 79%. This discrepancy can be due to the lower number of cases (n = 18) studied by Soller et al. [19] and/or to their use of nested PCR. Indeed, when we used nestedPCRin our samples, the frequency of the TMPRSS2-ERG transcript rose to 62%, but, additionally, three control samples (one HBP and two CP) also showed a clear positive signal (data not shown). False-positive results with highly sensitive PCR techniques for the detection of fusion transcripts are relatively rare, but are a major concern in patients with hematologic malignancies [20,21]. These can be due to contamination from previous positive cases or to the presence of very rare normal cells with abnormal molecular rearrangements characteristic of specific types of hematologic malignancies (e.g., BCR-ABL gene fusion in chronic myeloid leukemia and BCL2 gene rearrangement in follicular lymphoma) [20,21]. As a consequence, one should be aware of the potential risks of using nested PCR in diagnostic samples, despite scrupulous precautions taken to minimize contamination. Conversely, TMPRSS2-ETV1 fusion was not detected in any of our cases. This is in agreement with previous reported frequencies of 3% (1 of 32) and 0% (0 of 18) [5,18], suggesting that this molecular rearrangement, as with TMPRSS2-ETV4 fusion [6], is not a frequent event in prostate cancer.
To the best of our knowledge, this is the first study to report on the presence of TMPRSS2-ERG fusion in HGPIN lesions (21% of cases in our series). The detection of the TMPRSS2-ERG fusion gene in HGPIN is not unexpected because this lesion is considered to be a precursor of, at least, some prostate carcinomas [22]. Indeed, histologic data seem to support this hypothesis: HGPIN consists of architecturally benign prostatic acini lined by cells that seem to be malignant (Figure 2), and prostate carcinomas may have zones of HGPIN from which glands of carcinoma seem to stem [22–24]. In addition, prostates with carcinoma have more of these foci than do those without carcinoma; prostate glands with extensive HGPIN also have more multifocal carcinomas; and HGPIN lesions preferentially develop in the peripheral zone of the prostate, which is the site of origin for most adenocarcinomas [22,25]. Interestingly, in two patients (cases 42 and 58), the fusion transcript was detected in the HGPIN lesion, but not in the PCa present in the same gland. This observation supports the hypothesis that prostate carcinogenesis may be a multicentric process, in which at least two independent pathogenetic pathways may coexist in the same prostate, leading to independent neoplasias with or without the involvement of the ETS pathway.
Figure 2.
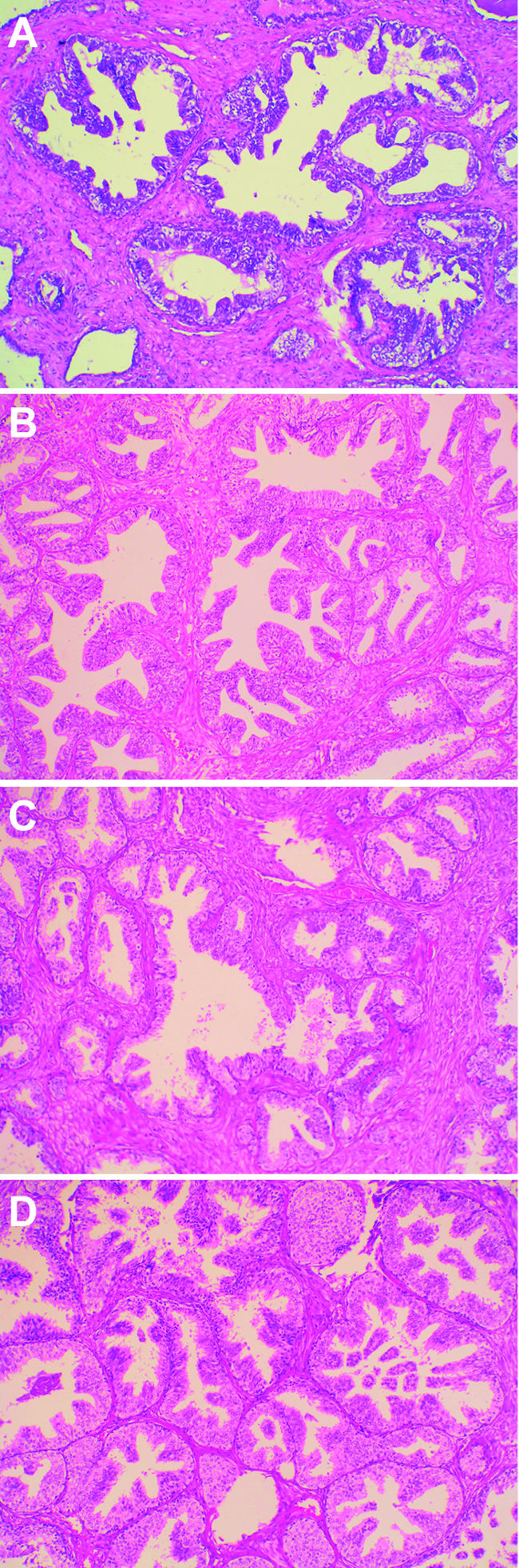
Representative images of four HGPIN lesions harboring the TMPRSS2-ERG fusion gene. (A) HGPIN 42. (B) HGPIN 58. (C) HGPIN 60. (D) HGPIN 83 (hematoxylin-eosin stain; original magnification, x25).
As opposed to prostate carcinomas with TMPRSS2-ERG fusion, where a clear association between TMPRSS2-ERG positivity and ERG overexpression was observed, this relationship was less constant in HGPIN lesions, with only one of four positive cases for the TMPRSS2-ERG fusion showing an ERG exons 5/6:exons 1/2 ratio above 2.5. These findings are compatible with the hypothesis that HGPIN lesions may initially be polyclonal proliferations, with the cells with TMPRSS2-ERG fusion being diluted in a pool of cells not harboring this alteration. Presumably, the HGPIN lesion may eventually be dominated by the clone with the TMPRSS2-ERG fusion as a result of clonal expansion, as shown by the detection of ERG overexpression in a subset of HGPIN lesions.
Although no correlation could be established between specific copy number changes detected by CGH and the presence of the TMPRSS2-ERG fusion transcript or ERG overexpression, several interesting conclusions can be drawn from our results. When we consider the group of PCa and HGPIN samples with TMPRSS2-ERG fusion, only eight of PCa cases and none of HGPIN samples showed copy number changes by CGH, indicating that the TMPRSS2-ERG fusion is an early pathogenetic event in prostate carcinogenesis that precedes the acquisition of chromosome imbalances. Because none of the TMPRSS2-ERG-positive cases showed a gain of 8q, a genomic imbalance recently identified as an independent predictor of poor survival [16], it is possible that the TMPRSS2-ERG gene fusion could represent a class of clinically less aggressive prostate carcinomas, but this needs to be addressed in a larger series.
In summary, we confirm that fusion of the ETS transcription factor ERG with the TMPRSS2 gene is a frequent event in prostate cancer. Furthermore, we demonstrate for the first time that TMPRSS2-ERG fusion can already be detected in a proportion of HGPIN lesions and that this molecular rearrangement is an early event that may precede chromosome-level alterations in prostate carcinogenesis.
Abbreviations
- HGPIN
high-grade prostatic intraepithelial neoplasia
- PCa
prostate adenocarcinomas
- ETS
erythroblast transformation specific
- CGH
comparative genomic hybridization
- RT-PCR
reverse transcriptase-polymerase chain reaction
Footnotes
This research was supported by research grant POCTI/SAU-OBS/58357/2004 from the Fundaçaäo para a Ciência e a Tecnologia.
References
- 1.Rowley JD. Chromosome translocations: dangerous liaisons revisited. Nat Rev Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 2.Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res. 2000;462:247–253. doi: 10.1016/s1383-5742(00)00006-5. [DOI] [PubMed] [Google Scholar]
- 3.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 4.Mitelman F, Mertens F, Johansson B. Prevalence estimates of recurrent balanced cytogenetic aberrations and gene fusions in unselected patients with neoplastic disorders. Genes Chromosomes Cancer. 2005;43:350–366. doi: 10.1002/gcc.20212. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 6.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 7.Hooper JD, Clements JA, Quigley JP, Antalis TM. Type II transmembrane serine proteases. Insights into an emerging class of cell surface proteolytic enzymes. J Biol Chem. 2001;276:857–860. doi: 10.1074/jbc.R000020200. [DOI] [PubMed] [Google Scholar]
- 8.Netzel-Arnett S, Hooper JD, Szabo R, Madison EL, Quigley JP, Bugge TH, Antalis TM. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev. 2003;22:237–258. doi: 10.1023/a:1023003616848. [DOI] [PubMed] [Google Scholar]
- 9.Vaarala MH, Porvari K, Kyllonen A, Lukkarinen O, Vihko P. The TMPRSS2 gene encoding transmembrane serine protease is overexpressed in a majority of prostate cancer patients: detection of mutated TMPRSS2 form in a case of aggressive disease. Int J Cancer. 2001;94:705–710. doi: 10.1002/ijc.1526. [DOI] [PubMed] [Google Scholar]
- 10.Afar DE, Vivanco I, Hubert RS, Kuo J, Chen E, Saffran DC, Raitano AB, Jakobovits A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61:1686–1692. [PubMed] [Google Scholar]
- 11.Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, Hood L, Nelson PS. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- 12.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–2478. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Foster CS, Bostwick DG, Bonkhoff H, Damber JE, van der Kwast T, Montironi R, Sakr WA. Cellular and molecular pathology of prostate cancer precursors. Scand J Urol Nephrol. 2000;205:19–43. doi: 10.1080/003655900750169284. [DOI] [PubMed] [Google Scholar]
- 14.Ribeiro FR, Diep CB, Jeronimo C, Henrique R, Lopes C, Eknaes M, Lingjaerde OC, Lothe RA, Teixeira MR. Statistical dissection of genetic pathways involved in prostate carcinogenesis. Genes Chromosomes Cancer. 2006;45:154–163. doi: 10.1002/gcc.20279. [DOI] [PubMed] [Google Scholar]
- 15.Kallioniemi OP, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D. Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer. 1994;10:231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]
- 16.Ribeiro FR, Jerónimo C, Henrique R, Fonseca D, Oliveira J, Lothe RA, Teixeira MR. 8q gain is an independent predictor of poor survival in diagnostic needle biopsies from prostate cancer suspects. Clin Cancer Res. 2006;12:3961–3970. doi: 10.1158/1078-0432.CCR-05-1977. [DOI] [PubMed] [Google Scholar]
- 17.Kirchhoff M, Gerdes T, Rose H, Maahr J, Ottesen AM, Lundsteen C. Detection of chromosomal gains and losses in comparative genomic hybridization analysis based on standard reference intervals. Cytometry. 1998;31:163–173. [PubMed] [Google Scholar]
- 18.ISCN, author. An International System for Human Cytogenetic Nomenclature Karger Landers. Basel, Switzerland: 1995. [Google Scholar]
- 19.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45:717–719. doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 20.Gleissner B, Rieder H, Thiel E, Fonatsch C, Janssen LA, Heinze B, Janssen JW, Schoch C, Goekbuget N, Maurer J, et al. Prospective BCR-ABL analysis by polymerase chain reaction (RT-PCR) in adult acute B-lineage lymphoblastic leukemia: reliability of RT-nested-PCR and comparison to cytogenetic data. Leukemia. 2001;15:1834–1840. doi: 10.1038/sj.leu.2402304. [DOI] [PubMed] [Google Scholar]
- 21.Bowman A, Jones D, Medeiros LJ, Luthra R. Quantitative PCR detection of t(14;18) bcl-2/JH fusion sequences in follicular lymphoma patients: comparison of peripheral blood and bone marrow aspirate samples. J Mol Diagn. 2004;6:396–400. doi: 10.1016/S1525-1578(10)60537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haggman MJ, Macoska JA, Wojno KJ, Oesterling JE. The relationship between prostatic intraepithelial neoplasia and prostate cancer: critical issues. J Urol. 1997;158:12–22. doi: 10.1097/00005392-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 23.McNeal JE. Origin and development of carcinoma in the prostate. Cancer. 1969;23:24–34. doi: 10.1002/1097-0142(196901)23:1<24::aid-cncr2820230103>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 24.McNeal JE, Bostwick DG. Intraductal dysplasia: a premalignant lesion of the prostate. Hum Pathol. 1986;17:64–71. doi: 10.1016/s0046-8177(86)80156-3. [DOI] [PubMed] [Google Scholar]
- 25.De Marzo AM, DeWeese TL, Platz EA, Meeker AK, Nakayama M, Epstein JI, Isaacs WB, Nelson WG. Pathological and molecular mechanisms of prostate carcinogenesis: implications for diagnosis, detection, prevention, and treatment. J Cell Biochem. 2004;91:459–477. doi: 10.1002/jcb.10747. [DOI] [PubMed] [Google Scholar]