Abstract
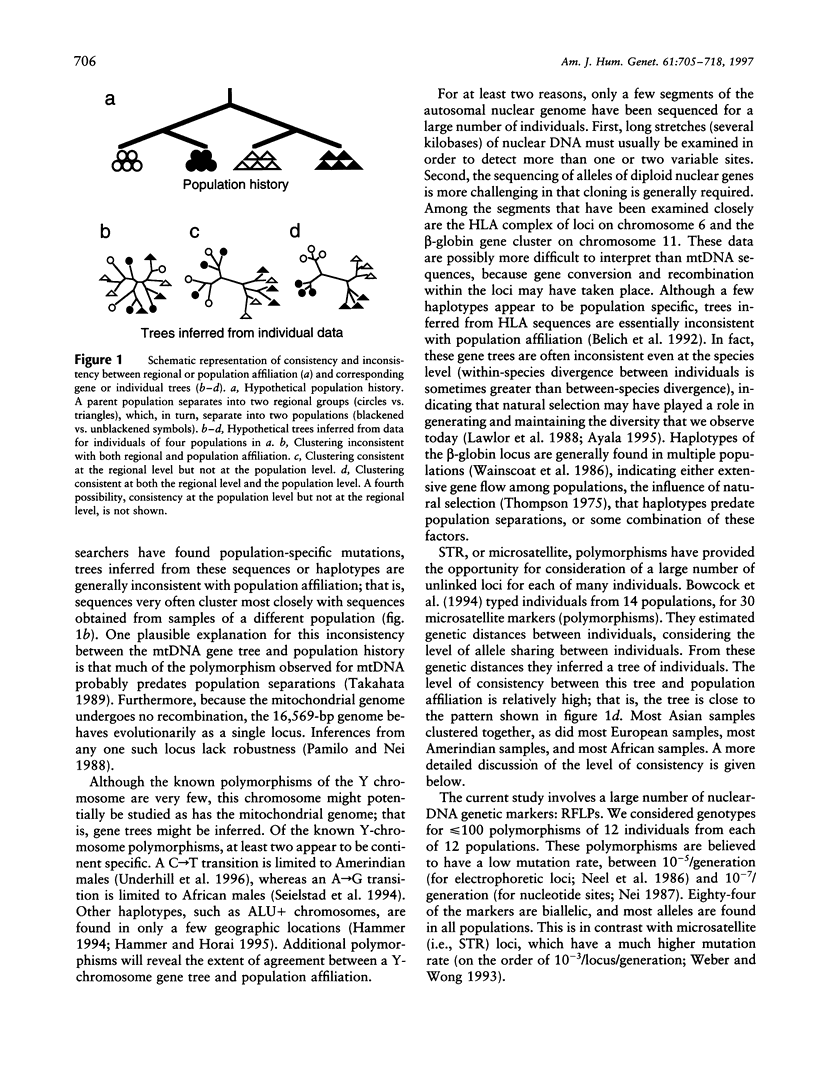
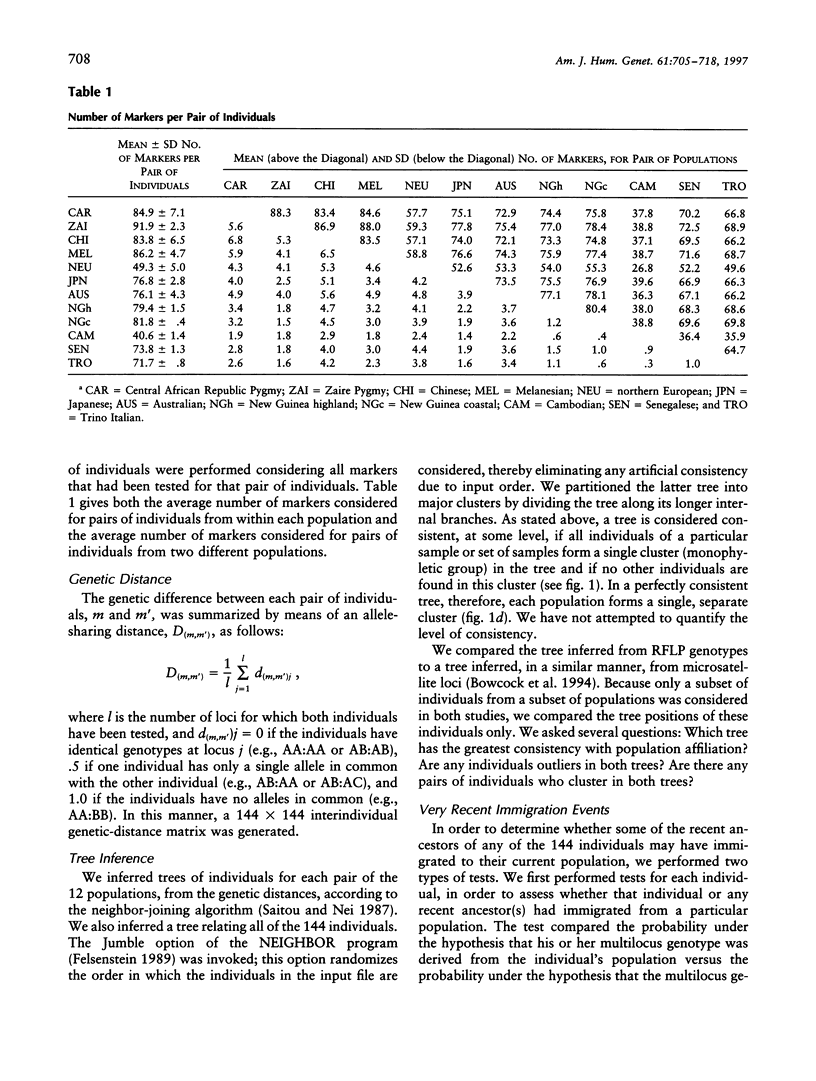
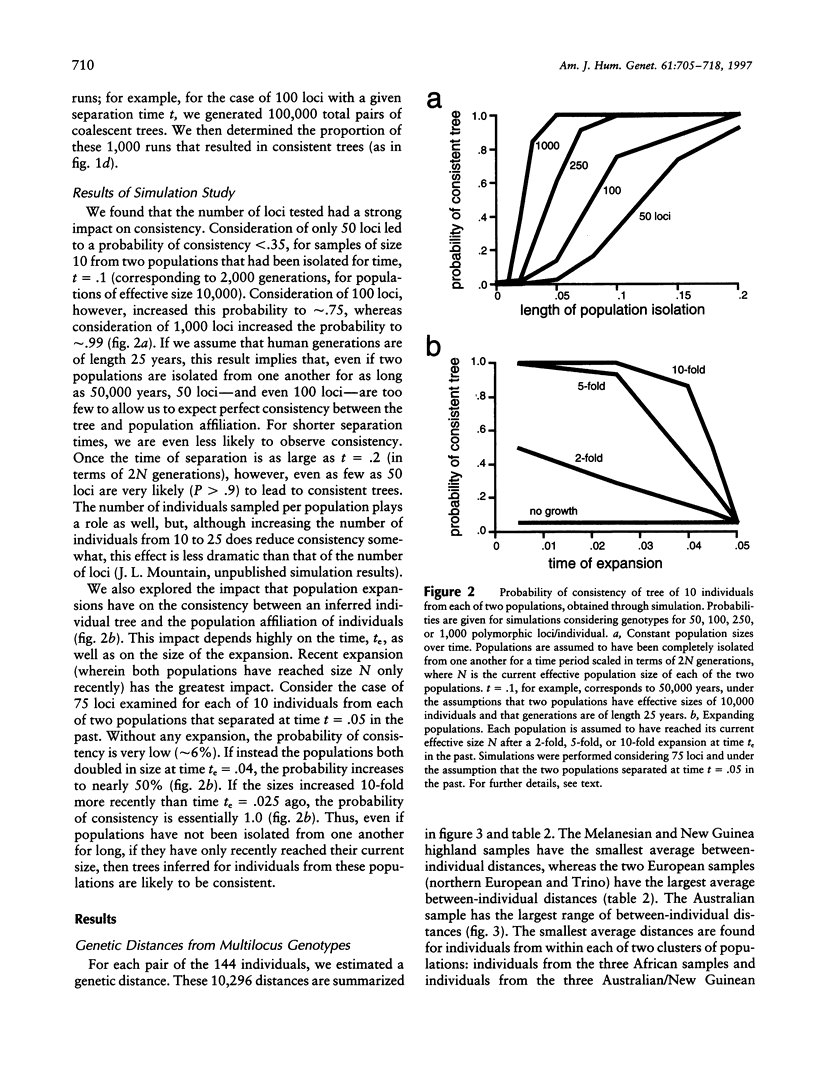
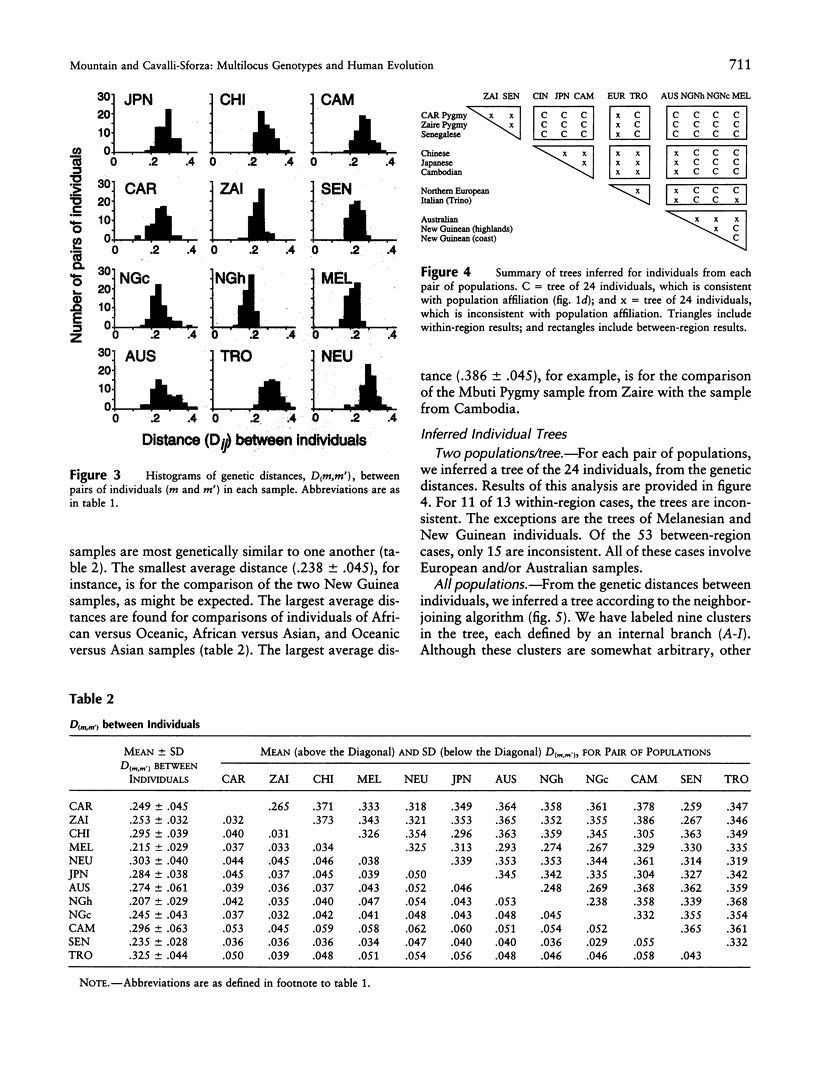
Our goal is to infer, from human genetic data, general patterns as well as details of human evolutionary history. Here we present the results of an analysis of genetic data at the level of the individual. A tree relating 144 individuals from 12 human groups of Africa, Asia, Europe, and Oceania, inferred from an average of 75 DNA polymorphisms/individual, is remarkable in that most individuals cluster with other members of their regional group. In order to interpret this tree, we consider the factors that influence the tree pattern, including the number of genetic loci examined, the length of population isolation, the sampling process, and the extent of gene flow among groups. Understanding the impact of these factors enables us to infer details of human evolutionary history that might otherwise remain undetected. Our analyses indicate that some recent ancestor(s) of each of a few of the individuals tested may have immigrated. In general, the populations within regional groups appear to have been isolated from one another for <25,000 years. Regional groups may have been isolated for somewhat longer.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson M. A., Gusella J. F. Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In Vitro. 1984 Nov;20(11):856–858. doi: 10.1007/BF02619631. [DOI] [PubMed] [Google Scholar]
- Aquadro C. F., Greenberg B. D. Human mitochondrial DNA variation and evolution: analysis of nucleotide sequences from seven individuals. Genetics. 1983 Feb;103(2):287–312. doi: 10.1093/genetics/103.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala F. J. The myth of Eve: molecular biology and human origins. Science. 1995 Dec 22;270(5244):1930–1936. doi: 10.1126/science.270.5244.1930. [DOI] [PubMed] [Google Scholar]
- Belich M. P., Madrigal J. A., Hildebrand W. H., Zemmour J., Williams R. C., Luz R., Petzl-Erler M. L., Parham P. Unusual HLA-B alleles in two tribes of Brazilian Indians. Nature. 1992 May 28;357(6376):326–329. doi: 10.1038/357326a0. [DOI] [PubMed] [Google Scholar]
- Bowcock A. M., Bucci C., Hebert J. M., Kidd J. R., Kidd K. K., Friedlaender J. S., Cavalli-Sforza L. L. Study of 47 DNA markers in five populations from four continents. Gene Geogr. 1987 Apr;1(1):47–64. [PubMed] [Google Scholar]
- Bowcock A. M., Hebert J. M., Mountain J. L., Kidd J. R., Rogers J., Kidd K. K., Cavalli-Sforza L. L. Study of an additional 58 DNA markers in five human populations from four continents. Gene Geogr. 1991 Dec;5(3):151–173. [PubMed] [Google Scholar]
- Bowcock A. M., Kidd J. R., Mountain J. L., Hebert J. M., Carotenuto L., Kidd K. K., Cavalli-Sforza L. L. Drift, admixture, and selection in human evolution: a study with DNA polymorphisms. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):839–843. doi: 10.1073/pnas.88.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock A. M., Ruiz-Linares A., Tomfohrde J., Minch E., Kidd J. R., Cavalli-Sforza L. L. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994 Mar 31;368(6470):455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Edwards A. W. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 1967 May;19(3 Pt 1):233–257. [PMC free article] [PubMed] [Google Scholar]
- Hammer M. F. A recent insertion of an alu element on the Y chromosome is a useful marker for human population studies. Mol Biol Evol. 1994 Sep;11(5):749–761. doi: 10.1093/oxfordjournals.molbev.a040155. [DOI] [PubMed] [Google Scholar]
- Hammer M. F., Horai S. Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet. 1995 Apr;56(4):951–962. [PMC free article] [PubMed] [Google Scholar]
- Johnson M. J., Wallace D. C., Ferris S. D., Rattazzi M. C., Cavalli-Sforza L. L. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns. J Mol Evol. 1983;19(3-4):255–271. doi: 10.1007/BF02099973. [DOI] [PubMed] [Google Scholar]
- Lawlor D. A., Ward F. E., Ennis P. D., Jackson A. P., Parham P. HLA-A and B polymorphisms predate the divergence of humans and chimpanzees. Nature. 1988 Sep 15;335(6187):268–271. doi: 10.1038/335268a0. [DOI] [PubMed] [Google Scholar]
- Lin A. A., Hebert J. M., Mountain J. L., Cavalli-Sforza L. L. Comparison of 79 DNA polymorphisms tested in Australians, Japanese and Papua New Guineans with those of five other human populations. Gene Geogr. 1994 Dec;8(3):191–214. [PubMed] [Google Scholar]
- Matullo G., Griffo R. M., Mountain J. L., Piazza A., Cavalli-Sforza L. L. RFLP analysis on a sample from northern Italy. Gene Geogr. 1994 Apr;8(1):25–34. [PubMed] [Google Scholar]
- Mountain J. L., Cavalli-Sforza L. L. Inference of human evolution through cladistic analysis of nuclear DNA restriction polymorphisms. Proc Natl Acad Sci U S A. 1994 Jul 5;91(14):6515–6519. doi: 10.1073/pnas.91.14.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel J. V., Satoh C., Goriki K., Fujita M., Takahashi N., Asakawa J., Hazama R. The rate with which spontaneous mutation alters the electrophoretic mobility of polypeptides. Proc Natl Acad Sci U S A. 1986 Jan;83(2):389–393. doi: 10.1073/pnas.83.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Roychoudhury A. K. Genic variation within and between the three major races of man, Caucasoids, Negroids, and Mongoloids. Am J Hum Genet. 1974 Jul;26(4):421–443. [PMC free article] [PubMed] [Google Scholar]
- Pamilo P., Nei M. Relationships between gene trees and species trees. Mol Biol Evol. 1988 Sep;5(5):568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- Poloni E. S., Excoffier L., Mountain J. L., Langaney A., Cavalli-Sforza L. L. Nuclear DNA polymorphism in a Mandenka population from Senegal: comparison with eight other human populations. Ann Hum Genet. 1995 Jan;59(Pt 1):43–61. doi: 10.1111/j.1469-1809.1995.tb01605.x. [DOI] [PubMed] [Google Scholar]
- Roberts-Thomson J. M., Martinson J. J., Norwich J. T., Harding R. M., Clegg J. B., Boettcher B. An ancient common origin of aboriginal Australians and New Guinea highlanders is supported by alpha-globin haplotype analysis. Am J Hum Genet. 1996 May;58(5):1017–1024. [PMC free article] [PubMed] [Google Scholar]
- Rogers A. R., Jorde L. B. Ascertainment bias in estimates of average heterozygosity. Am J Hum Genet. 1996 May;58(5):1033–1041. [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Seielstad M. T., Hebert J. M., Lin A. A., Underhill P. A., Ibrahim M., Vollrath D., Cavalli-Sforza L. L. Construction of human Y-chromosomal haplotypes using a new polymorphic A to G transition. Hum Mol Genet. 1994 Dec;3(12):2159–2161. doi: 10.1093/hmg/3.12.2159. [DOI] [PubMed] [Google Scholar]
- Shields G. F., Schmiechen A. M., Frazier B. L., Redd A., Voevoda M. I., Reed J. K., Ward R. H. mtDNA sequences suggest a recent evolutionary divergence for Beringian and northern North American populations. Am J Hum Genet. 1993 Sep;53(3):549–562. [PMC free article] [PubMed] [Google Scholar]
- Shriver M. D., Smith M. W., Jin L., Marcini A., Akey J. M., Deka R., Ferrell R. E. Ethnic-affiliation estimation by use of population-specific DNA markers. Am J Hum Genet. 1997 Apr;60(4):957–964. [PMC free article] [PubMed] [Google Scholar]
- Stoneking M., Jorde L. B., Bhatia K., Wilson A. C. Geographic variation in human mitochondrial DNA from Papua New Guinea. Genetics. 1990 Mar;124(3):717–733. doi: 10.1093/genetics/124.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N. Gene genealogy in three related populations: consistency probability between gene and population trees. Genetics. 1989 Aug;122(4):957–966. doi: 10.1093/genetics/122.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiercy J. M., Sanchez-Mazas A., Excoffier L., Shi-Isaac X., Jeannet M., Mach B., Langaney A. HLA-DR polymorphism in a Senegalese Mandenka population: DNA oligotyping and population genetics of DRB1 specificities. Am J Hum Genet. 1992 Sep;51(3):592–608. [PMC free article] [PubMed] [Google Scholar]
- Underhill P. A., Jin L., Zemans R., Oefner P. J., Cavalli-Sforza L. L. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci U S A. 1996 Jan 9;93(1):196–200. doi: 10.1073/pnas.93.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigilant L., Pennington R., Harpending H., Kocher T. D., Wilson A. C. Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9350–9354. doi: 10.1073/pnas.86.23.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainscoat J. S., Hill A. V., Boyce A. L., Flint J., Hernandez M., Thein S. L., Old J. M., Lynch J. R., Falusi A. G., Weatherall D. J. Evolutionary relationships of human populations from an analysis of nuclear DNA polymorphisms. Nature. 1986 Feb 6;319(6053):491–493. doi: 10.1038/319491a0. [DOI] [PubMed] [Google Scholar]
- Weber J. L., Wong C. Mutation of human short tandem repeats. Hum Mol Genet. 1993 Aug;2(8):1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Wijsman E. M. Techniques for estimating genetic admixture and applications to the problem of the origin of the Icelanders and the Ashkenazi Jews. Hum Genet. 1984;67(4):441–448. doi: 10.1007/BF00291407. [DOI] [PubMed] [Google Scholar]



