Abstract
T lymphoma invasion and metastasis 1 (Tiam1) is a metastasis-related gene of T lymphoma that is also involved in the metastasis of a variety of other cancers. In this study, we tested the hypothesis that Tiam1 is a determinant of proliferation and metastasis in colorectal cancer, and we examined the effect of the inhibition of Tiam1 expression on proliferation and metastasis. We succeeded in establishing the Tiam1 knockdown colorectal cancer cell line using human immunodeficiency virus lentivirus-mediated RNA interference (RNAi) and found that the silencing of Tiam1 resulted in the effective inhibition of in vitro cell growth and of the invasive ability of colorectal cancer cells. Using an orthotopic xenograft model in nude mice, we confirmed that Tiam1 silencing could reduce tumor growth by subcutaneous injection and could suppress lung and liver metastases of colorectal cancer cells. Our results suggest that Tiam1 truly plays a causal role in the metastasis of colorectal cancer and that RNAi-mediated silencing of Tiam1 may provide an opportunity to develop a new treatment strategy for colorectal cancer.
Keywords: Tiam1 gene, RNA interference, colorectal cancer, proliferation, metastasis
Introduction
Small guanosine triphosphate (GTP)-binding proteins of the Ras superfamily function as molecular switches in fundamental events such as signal transduction, cytoskeleton dynamics, and intracellular trafficking [1,2]. Mutations or aberrant regulation of these proteins can contribute to malignant phenotypes of human tumors. Guanine nucleotide exchange factors (GEFs), which catalyze the dissociation of guanosine diphosphate from inactive GTP-binding proteins, positively regulate these GTP-binding proteins in response to a variety of signals. Currently, many GEFs, including Vav1, LARG, Bcr, and T lymphoma invasion and metastasis 1 (Tiam1), have been identified as oncogenes [3–6].
Tiam1 was originally identified as an invasion-inducing and metastasis-inducing gene by proviral tagging, in combination with in vitro selection for invasiveness in T lymphoma cells [7]. The role of Tiam1 in cellular migration, invasion, and metastasis may not be limited to T lymphoma. It has been reported to be important in promoting tumor progression in a variety of cancers, such as breast cancer, colorectal cancer, lung cancer, and Ras-induced skin tumors [8–11].
In our previous studies, to screen metastasis-associated genes, we prepared a cDNA microarray of tumor metastasis-associated genes and obtained 51 genes that were closely associated with metastases of colorectal cancer [12,13]. Mining microarray gene expression data by literature profiling, we found that Tiam1 had potential relation to metastatic colorectal cancer [14]. To further investigate the effect of Tiam1 on metastases of colorectal cancer, we detected Tiam1 expression in cell lines and in different tissue specimens of colorectal cancer. We found that Tiam1 expression was highly related to metastatic potential in colorectal cancer and that Tiam1 was a metastasis-related gene [15]. However, it is unclear whether the knockdown of Tiam1 is responsible for metastatic phenotypes in colorectal cancer, and its mechanism is unclear as well.
In this study, we established a stable Tiam1 silencing colorectal cancer cell line with lentiviral vector-mediated RNA interference (RNAi) technology and determined whether Tiam1 silencing could influence in vitro proliferation and invasion and in vivo tumor growth and metastasis.
Materials and Methods
Cell Line and Animals
The human colorectal cancer cell line SW480/EGFP was established from SW480 by transfection of pEGFP-N1 plasmid and was cultured in RPMI 1640 medium (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin/streptomycin in a 5% CO2 humidified atmosphere at 37°C. The 293FT cell line, which stably expresses SV40 large T antigen and facilitates the optimal production of viruses, was cultured in Dulbecco's modified Eagle's medium (Gibco).
Four- to 6-week-old female/male athymic BALB/c nu/nu mice were purchased from the Central Laboratory of Animal Science at Southern University (Guangzhou, China) and maintained in laminar-flow cabinets under specific pathogen-free conditions.
Preparation of Lentiviral Vectors
Firstly, we selected four different sequences for targeting the Tiam1 gene using the BLOCK-It RNAi Designer (Invitrogen, Carlsbad, CA) (http://rnaidesigner.invitrogen.com/rnaiexpress/rnaiExpress.jsp) (Table 1). The preparation of lentiviral vectors expressing human Tiam1 short hairpin RNA (shRNA) was performed using the BLOCK-It Lentiviral RNAi Expression System (catalog no. K4944-00; Invitrogen), following the manufacturer's instruction. In brief, four pLenti6/Tiam1 expression vectors containing the human Tiam1 shRNA-expressing cassette were constructed. Replication-incompetent lentivirus was produced by cotransfection of the pLenti6/Tiam1 expression vector and ViraPower packaging mix (Invitrogen) containing an optimized mixture of three packaging plasmids: pLP1, pLP2, and pLP/VSVG) into 293FT cells. RNA; expression system includes ViraPower packaging mix. Viral supernatant was harvested 48 hours after transfection, filtered through a 0.45-µm cellulose acetate filter, and frozen at -70°C. The lentivirus containing the human Lamin A/C shRNA-expressing cassette (sequence 5′-CTGGACTTCCAGAAGAACA-3′) was used as a positive control for lentivirus production, and the lentivirus-only-containing pLenti6/U6 mock vector was used as negative control. Viral concentrations were determined by in vitro transduction and blasticidin selection.
Table 1.
Targeted Sequences of Tiam1 Gene for RNAi.
| Site | Start | Target sequence |
| A | 920 | 5′-GCATTCCTATACATCCAATGG-3′ |
| B | 2223 | 5′-GGTGAAATGCAGCTGTCTTCA-3′ |
| C | 3631 | 5′-GCACCTACGTGAAGGATTTAA-3′ |
| D | 4368 | 5′-GCTGTGGTCCTTGTGTATAAA-3′ |
Construction of Stable Silencing Lines
SW480/EGFP cells were transduced with specific or negative control lentiviral vectors and selected for stable integrants by culturing a complete medium containing blasticidin. After 12 days of selection, there were no viable cells in mock wells and discrete blasticidin resistance colonies.
Real-Time Reverse Transcription Polymerase Chain Reaction (PCR) Analysis for Tiam1
cDNA was synthesized by oligo dT primed reverse transcription from 2 µg of total RNA using an access reverse transcription system (Promega, Madison, WI). Real-time PCR was performed using Mx3000P Real-Time PCR System (Stratagene, La Jolla, CA) and Brilliant SYBR Green QPCR Master Mix kit (Stratagene), following the manufacturer's protocol. In brief, the reaction mixture (total volume, 25 µl) contained 500 ng of cDNA, the forward primer 5′-AAGACGTACTCAGGCCATGTCC-3′, and the reverse primer 5′-GACCCAAATGTCGCAGTCAG-3′ to amplify human Tiam1 (GeneBank, NM_003253) at a final concentration of 250 nM and with 12.5 µl of 2x SYBR Green QPCR Master Mix kit. Thermal cycling conditions were as follows: 95°C for 5 minutes and 45 cycles at 95°C for 40 seconds, followed by 58°C for 40 seconds and 72°C for 40 seconds. Experiments were performed in triplicate in the same reaction. Human β-actin gene was amplified as internal control. Target genes and β-actin gene were amplified in the same reaction. Comparative quantification is determined using the 2-ΒΒCt method [16].
Western Blot Analysis
Cells were washed twice with cold phosphate-buffered saline (PBS) and lysed on ice in RIPA buffer [1x PBS, 1% NP40, 0.1% sodium dodecyl sulfate (SDS), 5 mM EDTA, 0.5% sodium deoxycholate, and 1 mM sodium orthovanadate] with protease inhibitors. Protein lysates were resolved on 6% SDS polyacrylamide gel, electrotransferred to polyvinylidene fluoride membranes (Immobilon P; Millipore, Bedford, MA), and blocked in 5% nonfat dry milk in Tris-buffered saline, pH 7.5 (100 mM NaCl, 50 mM Tris, and 0.1% Tween-20). Membranes were immunoblotted overnight at 4°C with anti-Tiam1 polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Lamin A/C antibody (Chemicon, Temecula, CA), and anti-β-actin antibody (Santa Cruz Biotechnology), followed by their respective horseradish peroxidase-conjugated secondary antibodies. Signals were detected by enhanced chemiluminescence (Pierce, Rockford, IL).
In Vitro Cell Growth Assay
The cells were prepared at a concentration of 1 x 104 cells/ml, respectively. Aliquots (100 µl) were dispensed into 96-well microtiter plates. The cells were incubated for 1, 2, 3, 4, 5, 6, and 7 days, respectively, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed by adding 20 µl of MTT (5 mg/ml; Promega) for 4 hours. When MTT incubation had been completed, supernatants were removed. One hundred fifty milliliters of dimethyl sulfoxide (Sigma, St. Louis, MO) was added to each well. Fifteen minutes later, the absorbance value (OD) of each well was measured with a microplate reader set at 570 nm. All experiments were performed in triplicate.
Plate Clone Formation Assay
About 1 x 102 cells were added to each well (3 cm in diameter) of a six-well culture plate, and each cell group contained three wells. After incubation at 37°C for 12 days, the cells were washed twice with PBS and stained with Giemsa solution. The number of colonies containing ≥ 50 cells was counted under a microscope [plate clone formation efficiency = (number of colonies / number of cells inoculated) x 100%].
Flow Cytometry Analysis for Cell Cycle
Cells were harvested at an exponential growth phase, and single-cell suspensions containing 1 x 106 cells were fixed with 70% alcohol. Cell cycle was monitored using propidium iodide (PI) staining of nuclei. The fluorescence of DNA-bound PI in cells was measured with a FACScan flow cytometer (BD Biosciences), and the results were analyzed with ModFit 3.0 software (Verity Software House, Topsham, ME).
In Vitro Invasion Assay
This assay was performed using the method of Albini et al. [17], with modifications. The cell invasion chamber (Chemicon) contains a polycarbonate membrane with an 8-µm pore size, over which a thin layer of ECMatrix (Chemicon) is dried. The extracellular matrix (ECM) layer occludes membrane pores, blocking noninvasive cells from migrating. To the top chamber was added warm serum-free medium to rehydrate the ECM layer for 2 hours at room temperature. Tumor cells in a serum-free medium (300 µl containing 1 x 105 cells) were added to the top chamber. The bottom chamber was prepared using 10% FBS as chemoattactant. After 24 hours of incubation, noninvasive cells were removed with a cotton swab. The cells that had migrated through the membrane and had stuck to the lower surface of the membrane were fixed with methanol and stained with hematoxylin. For quantification, the cells were counted under a microscope in five predetermined fields (original magnification, x200).
In Vivo Tumor Growth Assay
To evaluate in vivo tumor growth, 1 x 107 Tiam1 silencing cells and mock virus-transduced cells were each injected subcutaneously into the left flank and right flank of six nude mice. After subcutaneous injection of cells, fluorescence emitted by cells was collected and imaged through a whole-body green fluorescence protein (GFP) imaging system (Lighttools, Encinitas, CA). Using IPP5.0 software (Cybermetics Co., Silver Spring, MD), images were analyzed such that whole-body optical images visualized real-time tumor growth and tumor area was calculated.
In Vivo Metastasis Assays
The cells were first harvested by trypsinization, washed thrice with cold serum-free medium, and resuspended with serum-free medium. Nude mice were injected subcutaneously with 1 x 107 cells. After 10 days, the tumors were extirpated and washed twice with antibiotic-containing RPMI 1640 medium. Cancerous tissue was divided into small pieces, approximately 1 mm in diameter. Nude mice were anesthetized with Nembutal (Sinopharm Chemical Reagent Co., Shanghai, China) and sterilized with alcohol. For colon surgical orthotopic implantation (SOI) using the method described previously [18], a small midline incision was made, and the colocecal part of the intestine was exteriorized. A small area of the colonal serosa was removed, and a 1-mm3 tumor fragment per mouse was implanted. A 7-0 nylon surgical suture was used to penetrate tumor pieces and to suture them to the intestine wall. The intestine was returned to the abdominal cavity, and the abdominal wall was closed with a 6-0 silk surgical suture. Whole-body optical images visualized real-time primary tumor growth and the formation of metastatic lesions. Two months later, all mice were killed, individual organs were excised, and metastases were checked by hematoxylin-eosin (H&E) staining.
Statistical Analysis
SPSS 12.0 software (Abbott Laboratories, North Chicago, IL) was used for statistical analysis. Assay differences between in vitro cell growth and in vivo tumor growth were tested for statistical significance using analysis of variance (ANOVA) for factorial design. Plate clone formation assay and in vitro invasion assay were tested using one-way ANOVA. Differences were considered statistically significant when P < .05.
Results
Establishment of Tiam1 Silencing Colorectal Cancer Cell Line
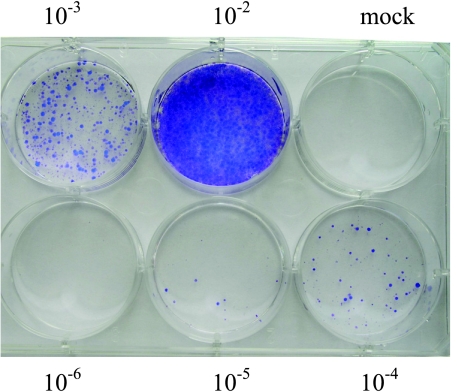
To deliver siRNA into the SW480/EGFP colorectal cancer cell line, we developed four lentiviral-based vectors that express different shRNA. After the cotransfection of pLenti6/Tiam1 vector and ViraPower packaging mix, replication-incompetent lentivirus was released into the medium and collected 48 hours after transfection. Viral stocks were tittered on SW480/EGFP cells, and 7 x 105 transducing units (TU)/ml was obtained (Figure 1).
Figure 1.
Titering lentiviral stock. Cells were either transduced with 10-fold serial dilutions of lentiviral supernatant (10-2-10-6 dilutions) or untransduced (Mock cells). Forty-eight hours posttransduction, the cells were placed under blasticidin selection. After 14 days of selection, the cells were stained with crystal violet, and colonies were counted. The virus titer was 7 x 105 TU/ml.
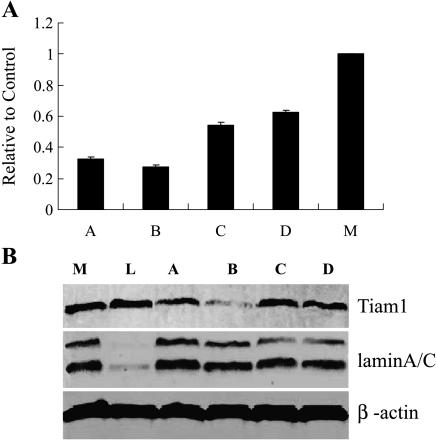
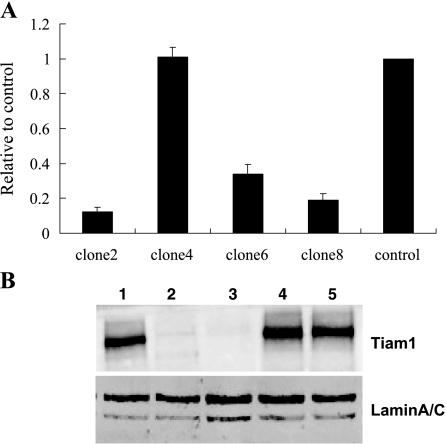
SW480/EGFP cells were transduced with pLenti6/U6, pLenti6/Tiam1, and pLenti6/Lamin A/C viral stocks and were selected by blasticidin 48 hours after transduction. Ten days posttransduction, cells were analyzed for Lamin A/C and Tiam1 expression using real-time PCR and Western blot analysis. As shown in Figure 2, site B lentiviral vector was most effective at blocking Tiam1 expression. There was no difference in cell morphology. Subsequently, we transduced pLenti6/Tiam1 (site B) and pLenti6/U6 lentiviruses into SW480/EGFP cells to select blasticidin-resistant single clones. Transduced clones were expanded and examined by real-time PCR and Western blot analysis (Figure 3). Results showed that Tiam1 knockdown clone 2 (Tiam1KD/clone 2), knockdown clone 8 (Tiam1KD/clone 8), and knockdown clone 6 (Tiam1KD/clone 6) exhibited 88%, 80%, and 66% reduction in Tiam1 protein, respectively. The clone transduced by pLenti6/U6 lentivirus exhibited no change in Tiam1 expression. For the sake of convenience, the SW480/EGFP cells and the clone transduced by pLenti6/U6 lentivirus are termed WTand Mock cells.
Figure 2.
Screening of the most effective targeting site for the Tiam1 gene by real-time PCR and Western blot analysis. (A) Quantification of Tiam1 mRNA expression in cells of different interference sites relative to controls (M, pLenti6/U6 Mock virus-transduced cells), as detected by real-time PCR. (B) Tiam1 protein expression in cells of different interference sites, as detected by Western blot analysis. Lane L shows that a greater degree of Lamin A/C knockdown was observed after transduction with a lentivirus containing the human Lamin A/C shRNA-expressing cassette. Lane B shows that lentiviral vectors were effective and that site B was the most effective construct.
Figure 3.
Confirmation of Tiam1 expression in different clones by real-time PCR and Western blot analysis. (A) Quantification of Tiam1 mRNA expression in clones transduced by the pLenti6/Tiam1 (site B) lentivirus relative to controls (clone transduced by pLenti6/U6 lentivirus). (B) Western blot analysis shows marked reduction of Tiam1 protein expression in clones 2 and 8. Lane 1, SW480/EGFP cells; lane 2, pLenti6/Tiam1 lentivirus-transduced clone 2; lane 3, pLenti6/Tiam1 lentivirus-transduced clone 8; lane 4, pLenti6/Tiam1 lentivirus-transduced clone 4; lane 5, pLenti6/U6 lentivirus-transduced clone.
Tiam1 Gene Silencing Suppresses Cell Proliferation In Vitro
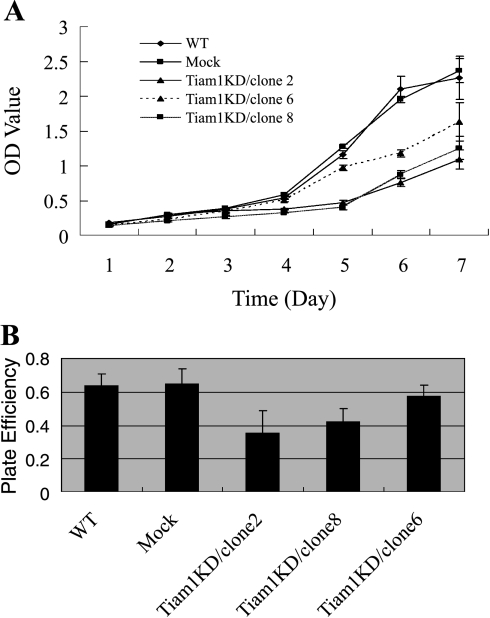
The effect of Tiam1 protein reduction on the proliferation of colorectal cancer cells was determined by MTT assay and plate clone formation assay. As shown in Figure 4A, although Mock cells showed in vitro growth ability approximating that of WT cells, Tiam1KD/clone 6 had reduced growth ability with respect to Mock cells, Tiam1KD/clone 8 had reduced growth ability with respect to Tiam1KD/clone 6, and Tiam1KD/clone 2 had reduced growth ability with respect to Tiam1KD/clone 8. Therefore, in vitro cell growth ability correlates with Tiam1 expression.
Figure 4.
Tiam1 gene silencing suppresses cell proliferation in vitro. (A) The in vitro proliferative abilities of WT cells, Mock cells, Tiam1KD/clone 2, Tiam1KD/clone 6, and Tiam1KD/clone 8 cells were evaluated by MTT assay. Each value represents the mean ± SD of the absorbance value (OD). Results showed that all Tiam1 knockdown cells grew significantly more slowly than WT and Mock cells, and Tiam1 protein expression correlated with cell proliferation. (B) The plate colony formation efficiency of WT cells, Mock cells, Tiam1KD/clone 2, Tiam1KD/clone 6, and Tiam1KD/clone 8 cells. Data represent the mean ± SD of triplicate dishes. Compared with WT and Mock cells, Tiam1KD/clone 2 had significantly reduced ability for colony formation (P < .01), and Tiam1KD/clone 8 also showed decreased clonogenicity (P < .05). Tiam1KD/clone 6, compared with WT and Mock cells, displayed no difference in clonogenicity (P > .05). Results showed that plate colony formation efficiency also correlated with Tiam1 protein expression.
The ability of cells to form colonies in plates was examined because there exists a correlation between clonogenicity and metastatic propensity. Figure 4B shows that Tiam1 knockdown cells, compared with WTand Mock cells, had a significant reduction in their ability to form colonies, and their ability to form colonies correlates with Tiam1 expression.
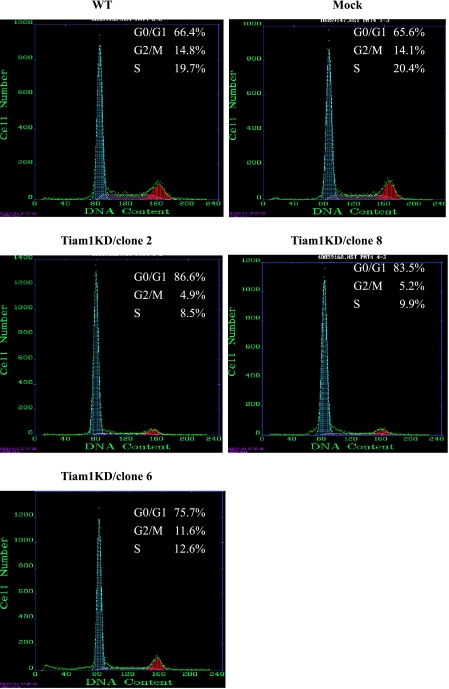
Flow Cytometry Analysis for Cell Cycle
In Figure 5, cell cycle analysis by flow cytometry reveals that the proportion of cells in the G0/G1, S, and G2/M phases for WT cells, Mock cells, Tiam1KD/clone 2, Tiam1KD/clone 6, and Tiam1KD/clone 8 cells were significantly different. The proportion of G0 to G1 phase was highest in Tiam1KD/clone 2, whereas the proportion of S phase was lowest in Tiam1KD/clone 2.
Figure 5.
Effect of Tiam1 knockdown on the cell cycle detected by flow cytometry analysis. Tiam1 silencing cells showed G0/G1 phase arrest and G2/M and S phase reduction. The difference was statistically significant.
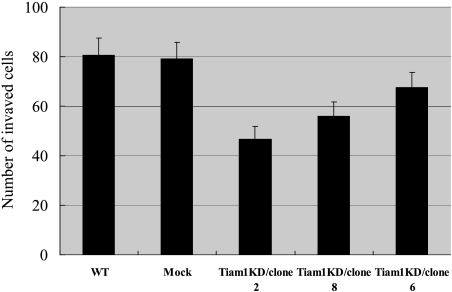
Tiam1 Gene Silencing Suppresses Cell Invasion In Vitro
Invasion through the ECM is an important step in tumor metastasis. ECMatrix serves as a reconstituted basement membrane matrix of proteins. The number of cells migrating to ECMatrix was counted. Tiam1 knockdown cells, compared with WT and Mock cells, display a remarkable decrease in invasiveness (P < .01). As shown in Figure 6, Tiam1KD/clone 2, Tiam1KD/clone 8, and Tiam1KD/clone 6 displayed a 1.72-fold, 1.44-fold, and 1.20-fold decrease in invasive ability, respectively, compared with WT cells. These results demonstrate that the invasive ability of colorectal cancer cells correlated with Tiam1 expression, and Tiam1 silencing alone is sufficient to attenuate invasion in colorectal cancer cells.
Figure 6.
Effect of Tiam1 knockdown on the invasive potential of colorectal cancer cells. In vitro invasion assay was carried out to compare and quantify the invasiveness of WT cells, Mock cells, Tiam1KD/clone 2, Tiam1KD/clone 6, and Tiam1KD/clone 8. Results are representative of three independent experiments, and bars represent the mean ± SD. Tiam1 silencing cells showed decreased invasion that correlated with Tiam1 protein expression.
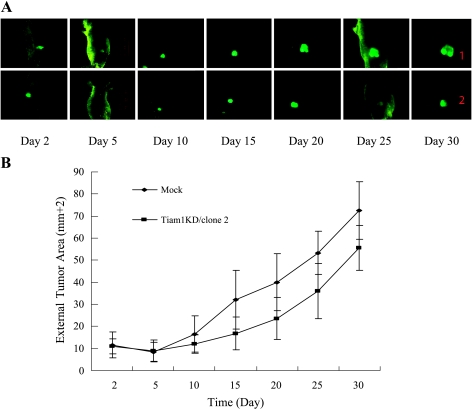
Tiam1 Gene Silencing Suppresses Cell Proliferation In Vivo
The effect of Tiam1 on in vivo tumor growth was assessed by the subcutaneous injection of Tiam1KD/clone 2 and Mock cells for 30 days. External fluorescent images can provide invaluable real-time data for tracking tumor growth. As shown in Figure 7, compared with that of Mock cells, the knockdown of Tiam1 expression progressed from a pronounced decrease in Tiam1KD/clone 2 cell growth by day 15 to a 1.3-fold decrease in tumor area by day 30 after cell injection.
Figure 7.
Tiam1 gene silencing suppresses cell proliferation in vivo. (A) Consecutive external whole-body fluorescence images of Mock cells and Tiam1KD/clone 2 tumors were obtained from days 2 to 30 after subcutaneous injection into nude mice. (B) Tumor areas were calculated by IPP5.0 software and were indicated as the mean ± SD of six mice. Compared with Mock cells, Tiam1KD/clone 2 had a significantly reduced in vivo proliferative ability (F = 2.256, P < .05).
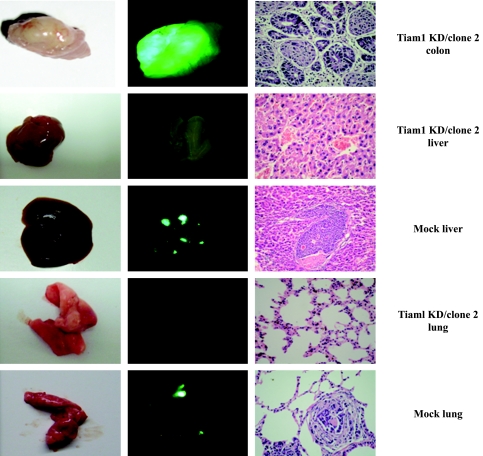
Tiam1 Silencing Reduces Metastatic Tumor Formation
To unambiguously elucidate the effect of Tiam1 on colorectal cancer metastasis, we performed in vivo metastasis assay by SOI. Mice were sacrificed 2 months later because the mice of the Mock group were moribund. Autopsy was performed, and the incidence of metastasis in the liver, lungs, and other organs was determined by macroscopic and histologic examinations (Figure 8). In the Mock group, 75% (six of eight) of mice developed peritoneal metastasis, which appeared as numerous bright green fluorescence on the peritoneum and on abdominal organs under a GFP imaging system. A comparison of external fluorescent images was made, and the results showed a direct image of mice and a external fluorescent image closely matching the direct image. In addition, the Tiam1KD/clone 2 in the orthotopical cecum of mice seldom expressed Tiam1 protein, whereas Mock cells in the orthotopical cecum of mouse stained positive for Tiam1 (Figure W1). In the Tiam1KD/clone 2 group, only 37.5% (three of eight) of animals had peritoneal metastasis. The incidence of hepatic metastasis and lung lesions in mice of the Mock group was 37.5% (three of eight) and 12.5% (one of eight), respectively, and metastatic lesions were validated by pathological methods. The Tiam1KD/clone 2 group did not produce detectable tumors in the liver and other organs. These results indicate that Tiam1 silencing was sufficient to decrease metastasis in colorectal cancer cells.
Figure 8.
Tiam1 silencing reduces metastatic lesions in vivo. We assessed the effect of Tiam1 silencing on metastasis using an orthotopic xenograft model in nude mice. After 2 months, the colon, lungs, and liver of mice were resected and analyzed for metastasis. Left panel: Direct image of colon, lungs, and liver. Middle panel: External fluorescent image of colon, lungs, and liver. Right panel: Histologic photomicrographs of colon, lung, and liver tissue sections stained with H&E (original magnification, x400).
Discussion
RNAi describes the phenomenon by which double-stranded RNA induces potent and specific inhibition of eukaryotic gene expression through the degradation of complementary messenger RNA (mRNA) and is functionally similar to the processes of posttranscriptional gene silencing. In the past few years, siRNA and shRNA have been widely used by researchers to silence the expression of many target genes because of their high specificity and apparent nontoxicity [19,20]. Furthermore, systems based on lentiviral vectors have provided new solutions to achieving stable shRNA-mediated knockdown [21,22].
In the present study, we used a lentivirus-mediated RNAi method to obtain an efficient knockdown of Tiam1 gene. Overexpression of Tiam1 seems to play a critical role in many human cancers. Recent results from other laboratories [9] and ours have demonstrated that increased Tiam1 expression correlates with the metastatic potential of human colorectal cancer and that Tiam1 was upregulated in the intestines of adenomatous polyposis coli (APC) conditional mutant mice [23]. In Tiam1-/- mice, susceptibility to the development of Ras-induced skin tumors, following application of a chemical carcinogenesis protocol, was significantly reduced [11]. This suggests that Tiam1 is a significant modulator of tumor development and metastasis, thus a potentially interesting therapeutic target.
In this study, we found that Tiam1 was one of the colorectal cancer genes associated with proliferation. The proliferation ability of tumor cells always correlates with metastatic phenotype and poor outcome [24]. We found that knockdown of Tiam1 expression strongly inhibited in vitro cell growth and colony formation efficiency. Moreover, through whole-body optical imaging that enables a continuous visual monitoring of malignant growth within intact animals, we found that silencing of Tiam1 expression appeared to have an inverse correlation with tumorigenicity. All data indicated that Tiam1 may be a positive regulator of tumor growth in colorectal cancer.
In agreement with Minard [9], our experiments demonstrated that knockdown of Tiam1 expression in colorectal cancer cells reduced their ability to invade ECMatrix-coated membranes in an invasion chamber assay. These results support the involvement of Tiam1 with the malignant behavior of human colorectal cancer cells and underline the relevance of a stable lentivirus-based Tiam1 knockdown mediated by RNAi in the impairment of the invasive ability of colorectal cancer.
Tiam1 has been extensively studied for the exchange factor's role in cellular migration, adhesion, and invasion. However, limited information on the promotion of in vivo metastasis is available. Our results demonstrated clearly that downregulated Tiam1 expression in colorectal cancer cells significantly suppressed the metastatic potential in Tiam1 silencing cells. We checked Tiam1 expression levels in Tiam1 knockdown clone before each experiment to ensure that Tiam1 expression was consistently of low level. We used an orthotopic xenograft model to evaluate the effect of Tiam1 silencing on metastasis because the models developed with SOI can exhibit patient-like metastasis. The reduced expression of Tiam1 in SW480/EGFP cells abrogated their ability to develop lung and hepatic metastases.
Taken together, our results indicate that the silencing of Tiam1 expression by RNAi suppressed the proliferation of colorectal cancer both in vitro and in vivo. This is the first study to demonstrate that targeting Tiam1 in vivo reduced the metastatic potential of colorectal cancer. Tiam1 is an important determinant of malignant cellular behavior and is a promising target for therapeutic intervention.
Supplementary Material
Acknowledgement
We thank Jimin Gao (Institute of Molecular Immunology, Southern Medical University, Guangzhou, China) for revising the manuscript.
Abbreviations
- Tiam1
T lymphoma invasion and metastasis 1
- RNAi
RNA interference
Footnotes
This work was supported by the National Natural Science Foundation of China (grants 30370649 and 30500242).
This article refers to supplementary material, which is designated by “W” (i.e., Figure W1) and is available online at www.bcdecker.com.
References
- 1.Haeusler LC, Blumenstein L, Stege P, Dvorsky R, Ahmadian MR. Comparative functional analysis of the Rac GTPases. FEBS Lett. 2003;555(3):556–560. doi: 10.1016/s0014-5793(03)01351-6. [DOI] [PubMed] [Google Scholar]
- 2.Michiels F, Collard JG. Rho-like GTPases: their role in cell adhesion and invasion. Biochem Soc Symp. 1999;65:125–146. [PubMed] [Google Scholar]
- 3.Denicola G, Tuveson DA. VAV1: a new target in pancreatic cancer? Cancer Biol Ther. 2005;4(5):509–511. doi: 10.4161/cbt.4.5.1781. [DOI] [PubMed] [Google Scholar]
- 4.Perrot V, Vazquez-Prado J, Gutkind JS. Plexin B regulates Rho through the guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. J Biol Chem. 2002;277(45):43115–43120. doi: 10.1074/jbc.M206005200. [DOI] [PubMed] [Google Scholar]
- 5.Bassermann F, Jahn T, Miething C, Seipel P, Bai RY, Coutinho S, Tybulewicz VL, Peschel C, Duyster J. Association of Bcr-Abl with the proto-oncogene Vav is implicated in activation of the Rac-1 pathway. J Biol Chem. 2002;277(14):12437–12445. doi: 10.1074/jbc.M112397200. [DOI] [PubMed] [Google Scholar]
- 6.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6(2):167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 7.Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, Stam JC, Berns A, Collard JG. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77(4):537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 8.Minard ME, Kim LS, Price JE, Gallick GE. The role of the guanine nucleotide exchange factor Tiam1 in cellular migration, invasion, adhesion and tumor progression. Breast Cancer Res Treat. 2004;84(1):21–32. doi: 10.1023/B:BREA.0000018421.31632.e6. [DOI] [PubMed] [Google Scholar]
- 9.Minard ME, Herynk MH, Collard JG, Gallick GE. The guanine nucleotide exchange factor Tiam1 increases colon carcinoma growth at metastatic sites in an orthotopic nude mouse model. Oncogene. 2005;24(15):2568–2573. doi: 10.1038/sj.onc.1208503. [DOI] [PubMed] [Google Scholar]
- 10.Hou M, Tan L, Wang X, Zhu YS. Antisense Tiam1 downregulates the invasiveness of 95D cells in vitro. Acta Biochim Biophys Sin (Shanghai) 2004;36(8):537–540. doi: 10.1093/abbs/36.8.537. [DOI] [PubMed] [Google Scholar]
- 11.Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417(6891):867–871. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- 12.Sun Q, Ding YQ, Gao XQ, Yan S, Han JX. Development and application of cDNA microarray of tumor metastasis-associated genes. Di Yi Jun Yi Da Xue Xue Bao. 2002;22(12):1070–1075. [PubMed] [Google Scholar]
- 13.Weng DS, Ding YQ, Sun Q. Differences in gene expression profile between high and low metastatic human colorectal cancer cell lines. Di Yi Jun Yi Da Xue Xue Bao. 2004;24(3):256–259. [PubMed] [Google Scholar]
- 14.Huang ZX, Sun Q, Ding YQ, Yao KT. Mining microarray gene expression data of metastatic colorectal cancer by literature profiling. Di Yi Jun Yi Da Xue Xue Bao. 2003;23(11):1195–1197. [PubMed] [Google Scholar]
- 15.Liu L, Wu DH, Ding YQ. Tiam1 gene expression and its significance in colorectal carcinoma. World J Gastroenterol. 2005;11(5):705–707. doi: 10.3748/wjg.v11.i5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, McEwan RN. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 1987;47(12):3239–3245. [PubMed] [Google Scholar]
- 18.Rashidi B, Gamagami R, Sasson A, Sun FX, Geller J, Moossa AR, Hoffman RM. An orthotopic mouse model of remetastasis of human colon cancer liver metastasis. Clin Cancer Res. 2000;6(6):2556–2561. [PubMed] [Google Scholar]
- 19.Gartel AL, Kandel ES. RNA interference in cancer. Biomol Eng. 2006;23(1):17–34. doi: 10.1016/j.bioeng.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Lu PY, Xie F, Woodle MC. In vivo application of RNA interference: from functional genomics to therapeutics. Adv Genet. 2005;54:117–142. doi: 10.1016/S0065-2660(05)54006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbas-Terki T, Blanco-Bose W, Deglon N, Pralong W, Aebischer P. Lentiviral-mediated RNA interference. Hum Gene Ther. 2002;13(18):2197–2201. doi: 10.1089/104303402320987888. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Rossi JJ. Lentiviral vector delivery of siRNA and shRNA encoding genes into cultured and primary hematopoietic cells. Methods Mol Biol. 2005;309:261–272. doi: 10.1385/1-59259-935-4:261. [DOI] [PubMed] [Google Scholar]
- 23.Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, Batlle E, Simon-Assmann P, Clevers H, Nathke IS, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18(12):1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6(2):99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.