Abstract
ECV is an E3 ubiquitin ligase complex, which is composed of elongins B and C, Rbx1, Cul2, and the substrate-conferring von Hippel-Lindau (VHL) tumorsuppressor protein that targets the catalytic α subunit of hypoxia-inducible factor (HIF) for oxygen-dependent ubiquitin-mediated destruction. Mutations in VHL that compromise proper HIFα regulation through ECV have been documented in the majority of renal cell carcinomas, underscoring the significance of the VHL-HIF pathway in renal epithelial oncogenesis. Recent evidence has shown that the modification of Cul2 by the ubiquitin-like molecule NEDD8 increases the activity of ECV to ubiquitylate HIFα. However, the underlying mechanism responsible for the NEDD8-mediated induction of ECV function is unknown. Here, we demonstrate that oxygen-dependent recognition of HIFα by VHL triggers Rbx1-dependent neddylation of Cul2, which preferentially engages the E2 ubiquitin-conjugating enzyme UbcH5a. These events establish a central role for the neddylation of Cul2 in a previously unrecognized, temporally coordinated activation of ECV with the recruitment of its substrate HIFα.
Keywords: Cul2, NEDD8, UbcH5a, HIFα, VHL
Introduction
Mutation of the von Hippel-Lindau (VHL) protein is the cause of VHL disease, which is characterized by the development of hypervascular tumors in multiple organs, including the central nervous system (cerebellum and spinal cord), retina, adrenal glands, and kidneys [1,2]. Tumor development is observed on the loss of the remaining wild-type VHL allele in a susceptible cell, thus conforming to Knudson's two-hit model. Biallelic inactivation of VHL locus is also associated with the majority (85%) of sporadic clear cell-renal cell carcinoma (the predominant form of kidney cancer) and also accounts for approximately 30% of sporadic cerebellar hemangioblastomas [2]. In addition, certain mutations in VHL cause congenital polycythemia [2].
VHL is a substrate-recognition component of an SCF (Skp1/Cdc53 or Cul1/F-box protein)-like E3 ubiquitin ligase complex, ECV, which is composed of elongins B and C, Cul2, and a RING finger protein Rbx1 (also known as ROC1 or Hrt1) that targets hypoxia-inducible factor (HIF) α subunits selectively under normal oxygen tension [3–6]. There are three HIFα genes (HIF1α, HIF2α, and HIF3α) in humans, and, under hypoxia, HIFα binds to β subunit [also known as aryl hydrocarbon receptor nuclear translocator (ARNT)] to form an active heterodimeric HIF transcription factor that upregulates the expression of numerous genes in response to compromised oxygen availability [7]. HIFα and ARNT are members of the basic helix-loop-helix (6HLH)-Per/ARNT/Sim (PAS) family of DNA-binding proteins. The basic domain is essential for binding DNA, and the HLH and PAS domains are required for heterodimerization and DNA binding [7]. HIFα contains two transactivation domains (TADs) located in the N-terminus (NAD) and C-terminus (CAD), whereas ARNTcontains just one TAD in the C-terminus. HIFα protein stability is conferred through the oxygen-dependent degradation (ODD) domain, which is targeted for ECV-dependent ubiquitylation upon hydroxylation of the conserved prolyl residue within the ODD by a class of prolyl hydroxylases (PHDs) in the presence of oxygen [7].
Unlike the a subunits, ARNT is constitutively expressed and stable. On hypoxia or inactivating mutations on VHL, HIFα escapes recognition by ECV and is consequently stabilized, recruits p300/CBP through CAD, and binds ARNT [7]. The active heterodimeric HIF complex binds to the hypoxia-responsive element (HRE) in enhancers/promoters to trigger the transcription of hypoxia-inducible genes that promote, in adaptation to hypoxia, angiogenesis, erythropoiesis, and glycolysis, as well as genes involved in iron metabolism and cell survival [7,8]. The overproduction of various HIF target gene products, such as vascular endothelial growth factor and erythropoietin, which are known to promote neovascularization and the production of oxygen-carrying red blood cells, respectively, likely explains the hypervascular nature of tumors, as well as polycythemia, associated with VHL disease.
The E3 function of both SCF and ECV is dependent on the recruitment of their E2 ubiquitin-conjugating enzymes, Cdc34 and UbcH5a, respectively [4,9–12]. Cullins are scaffold components of SCF and ECV ubiquitin ligases, and are covalently modified by the ubiquitin-like molecule NEDD8 [13]. NEDD8 is attached to target proteins in a manner analogous to ubiquitin attachment, involving the NEDD8-activating APP-BP1/Uba3 heterodimeric enzyme (NAE; E1) and the NEDD8-conjugating enzyme (NCE; E2) UbcH12 [14,15]. Importantly, the overall E3 ubiquitin ligase activity of yeast SCF is enhanced by covalent modification of the Cullin orthologue Cdc53 by the NEDD8 orthologue Rub1 [16,17]. Similarly, the activity of the human SCFβTrCP and SCFSkp2 complex is increased by the neddylation of Cul1, which facilitates the ubiquitylation of IκBα and p27, respectively [18–22]. In accordance, NEDD8 modification of Cul2 enhances the E3 activity of the SCF-like ECV complex in vivo [23].
In an effort to define the mechanisms that regulate SCF function, the core Cullin/Rbx1 complex was shown to be required for Cdc34 recruitment by the yeast SCF [24,25]. Wu et al. [26] subsequently demonstrated that the neddylated Cul1/Rbx1 complex was significantly better than unneddylated Cul1/Rbx1 in supporting the Cdc34-mediated assembly of polyubiquitin chains. In support, NEDD8 modification of Cul1 was shown to directly enhance the binding of ubiquitinconjugated E2 Ubc4 to SCFβTrCP [19]. In addition, p120CAND1 was identified to interact selectively with unneddylated Cul1 to cause Skp1 dissociation from the SCF complex. Conversely, neddylation of Cul1 prevented p120CAND1 binding, allowing SCF complex formation and activity [27–31].
Although these aforementioned studies provide several models that explain, in part, Cul1 and/or Cdc53 neddylation-mediated regulation of SCF activity, the mechanism by which Cul2 neddylation promotes ECV ubiquitin ligase function is entirely unknown. Here, we show that the NEDD8 modification of Cul2 requires Rbx1, which dramatically enhances binding to UbcH5a. ECV that has engaged HIF1α contains preferentially NEDD8-modified Cul2, whereas ECV consisting of mutant VHL incapable of binding HIFα exclusively associates with unmodified Cul2. In addition, increased HIF1α association with VHL results in a corresponding increase of Cul2 neddylation in the context of ECV. These results suggest that the oxygen-dependent recognition/binding of HIFα through VHL triggers Rbx1-mediated neddylation of Cul2, which promotes the engagement of UbcH5a to the ECV complex, thereby establishing a central role for the neddylation of Cul2 in the temporally coordinated activation of ECV with the recruitment of its substrate HIFα.
Materials and Methods
Cells
The HEK293A human embryonic kidney cell line (American Type Culture Collection, Rockville, MD) was maintained in Dulbecco's modified Eagle's medium containing 10% heatinactivated fetal bovine serum (Sigma-Aldrich, Oakville, Canada) at 37°C in a humidified atmosphere with 5% CO2.
Antibodies
Anti-HA (Y11), anti-myc polyclonal, and anti-His monoclonal antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-HA (12CA5), anti-HIF1α monoclonal, and anti-Cul2 polyclonal antibodies were obtained from Boehringer (Mannheim, Germany), Novus Biologicals (Littleton, CO), and Zymed (San Francisco, CA), respectively. MG132 proteasome inhibitor was obtained from Boston Biochem (Cambridge, MA).
Plasmids
The generation of mammalian expression plasmids pRCCMV-HA-Cul2(WT) and pRC-CMV-HA-Cul2(K689R) has been described previously [23], and pcDNA3.1-HACul2ΔC(1–600) was kindly provided by Dr. William G. Kaelin. pcDNA3.1-myc-Rbx1 was generously provided by Dr. James DeCaprio. pRC-CMV-HA-VHL(WT) and pcDNA3-HA-HIF1α have been described previously [5]. Purified His-UbcH5a was obtained from Boston Biochem.
Immunoprecipitation and Immunoblotting
Immunoprecipitation was performed as described previously [23]. In brief, cells were lysed in EBC buffer (50 mM Tris, pH 8.0; 120 mM NaCl; and 0.5% NP-40) and supplemented with protease and phosphatase inhibitors (Roche, Laval, Canada). Cell lysates were immunoprecipitated with indicated antibodies in the presence of protein A-Sepharose beads (Amersham Biosciences, Piscataway, NJ). Bound proteins were washed five times with NETN buffer (20 mM Tris, pH 8.0; 120 mM NaCl; 1 mM EDTA; and 0.5% NP-40), eluted by boiling in sodium dodecyl sulfate (SDS)-containing sample buffer, and resolved on SDS polyacrylamide gel electrophoresis (PAGE). Western blot analysis was performed as described previously [23].
In Vitro Binding Assays
In vitro translation in the presence of [35S]methionine was performed using the TNT reticulocyte in vitro transcription/translation system (Promega, Madison, WI), according to the manufacturer's instructions. Indicated translation products were mixed at 37°C for 60 minutes. Reaction mixtures were then incubated with indicated antibodies in the presence of protein A-Sepharose beads in 700 µl of EBC buffer (50 mM Tris, pH 8.0; 120 mM NaCl; and 0.5% NP-40) at 4°C for 1 hour. After five washes with NETN buffer (20 mM Tris, pH 8.0; 120 mM NaCl; 1 mM EDTA; and 0.5% NP-40), bound proteins were resolved on SDS-PAGE and visualized by autoradiography.
In Vitro Neddylation Assay
In vitro neddylation assay was performed as previously described [23].
Hypoxia Treatment of Cells
Cells were maintained at 1% O2 in a ThermoForma (Marietta, OH) hypoxia chamber. Cell lysates were prepared in the chamber in a hypoxic environment before further experimentation.
Results and Discussion
Rub1 covalently modifies Cdc53 to modulate the activity of SCF in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Arabidopsis thaliana [16,17,32,33]. Likewise, the mammalian Rub1 orthologue NEDD8 covalently associates with Cullins [13]. The impairment of Cul1 neddylation reduces the ubiquitylation of the known SCF substrates p27 and IκBα [21,22]. We have previously shown that Cul2 of the ECV complex is modified by NEDD8 under physiological conditions and that the attenuation of this process is associated with a decreased ability of ECV to polyubiquitylate HIFα [23]. However, the precise nature of how the NEDD8 conjugation of Cul2 modulates the activity of ECV is unknown.
Neddylation Status of Cul2 Does Not Influence Binding to Rbx1
Rbx1 has been shown to be involved in the recruitment of E2 ubiquitin-conjugating enzyme [24,25]. The binding of Rbx1 has been mapped outside the Cul1 neddylation site [18]. However, the Rbx1-binding region on Cul2 has not been established, and it is unclear whether neddylation of Cul2 would influence binding to Rbx1 by promoting subtle conformational changes to Cul2. Thus, we asked whether the neddylation of Cul2 influenced the engagement of Rbx1 as a potential mechanism that promotes the activity of ECV. 35S-labeled in vitro-translated HA-Cul2(WT) or a neddylationdefective HA-Cul2(K689R) mutant [23] was mixed with 35S-labeled in vitro-translated myc-Rbx1, immunoprecipitated with an anti-HA antibody, resolved on SDS-PAGE, and visualized by autoradiography (Figure 1A). As previously demonstrated [23], HA-Cul2(WT) migrated as a doublet, where the more slowly migrating protein represents Cul2 covalently linked to NEDD8. As expected and as previously shown [23], HA-Cul2(K689R) failed to be neddylated and therefore migrated as a single band. However, Rbx1 coprecipitated with both HA-Cul2(WT) and HA-Cul2(K689R) (Figure 1A). An analogous experiment was performed in vivo, where human embryonic kidney HEK293A cells were transiently transfected with plasmids encoding HA-Cul2(WT) or HA-Cul2(K689R) with or without a plasmid encoding myc-Rbx1. Cells were lysed, immunoprecipitated with an anti-HA antibody, resolved by SDS-PAGE, and immunoblotted with an anti-HA (upper panel) or anti-myc (lower panel) antibody (Figure 1B). Consistent with in vitro data, similar levels of myc-Rbx1 coprecipitated with either neddylatable HACul2( WT) or unneddylatable HA-Cul2(K689R), suggesting that the neddylation status/capability of Cul2 does not influence its interaction with Rbx1 (Figure 1B).
Figure 1.
Cul2 neddylation status does not influence association with Rbx1. (A) 35S-labeled in vitro-translated HA-Cul2(WT) or neddylation-defective HACul2(K689R) mutant was, where indicated, mixed with 35S-labeled in vitro-translated myc-Rbx1 and immunoprecipitated with indicated antibodies. Bound proteins were resolved by SDS-PAGE and visualized by autoradiography. (B) HEK293A cells were transiently transfected with mammalian expression plasmids encoding myc-Rbx1, HA-Cul2(WT), or HA-Cul2(K689R), alone or in combination. Cells were lysed and immunoprecipitated with indicated antibodies. Bound proteins were resolved by SDS-PAGE and immunoblotted with anti-Cul2 (upper panel) and anti-myc (lower panel) antibodies. IP = immunoprecipitation.
Rbx1 Acts as an E3 NEDD8 Ligase to Promote the Neddylation of Cul2
Interestingly, the ectopic expression of Rbx1 promoted the accumulation, albeit modest, of NEDD8-modified Cul2 (Figure 1B, cf. lanes 3 and 4). We asked whether Rbx1 promoted Cul2 neddylation by performing an in vitro Cul2 neddylation assay, as previously described [23]. In vitro-translated HA-Cul2(WT), in the absence of neddylation reaction, produced minimal modification of Cul2 by NEDD8 (Figure 2A, lane 1), whereas in the presence of neddylation reaction (containing purified NAE, UbcH12, NEDD8, and ATP; see the Materials and Methods section), a greater fraction of HA-Cul2 became neddylated. The addition of exogenous in vitro-translated Rbx1 to this reaction significantly shifted the status of Cul2 toward NEDD8-modified Cul2 (Figure 2A, lane 3). The slight modification of Cul2 observed in the absence of exogenous Rbx1 was likely due to the presence of endogenous Rbx1 in the rabbit reticulocyte lysate used in the in vitro translation reaction (Figure 2A, lane 1). In a complementary experiment, we made use of the Cul2ΔC mutant, which is a C-terminal Cul2 truncation mutant (1–600) capable of binding Rbx1 but lacking a NEDD8 conjugation site (Figure 2B). The incubation of co-in vitro-translated HA-Cul2/myc-Rbx1 with increasing amounts of Cul2ΔC before neddylation reaction resulted in the attenuation of Cul2 neddylation (Figure 2C, left panel). In addition, the incubation of in vitro-translated myc-Rbx1 with increasing amounts of Cul2ΔC before the in vitro Cul2 neddylation reaction resulted in a similar diminution of Cul2 neddylation (Figure 2C, right panel). These results, taken together, suggest that Rbx1 functions as an E3 NEDD8 ligase to direct the specific targeting of Cul2 for neddylation in concert with NAE and NCE/UbcH12.
Figure 2.
Rbx1 promotes Cul2 neddylation. (A) HA-Cul2(WT) was translated in vitro alone (lanes 1 and 2) or cotranslated with myc-Rbx1 (lane 3) in the presence of [35S]methionine and subjected to a neddylation reaction, where indicated. Reaction mixtures were subsequently immunoprecipitated with anti-HA antibody, resolved on SDS-PAGE, and visualized by autoradiography. (B) myc-Rbx1, in combination with HA-Cul2ΔC, was translated in vitro in the presence of [35S]methionine and immunoprecipitated with indicated antibodies. Bound proteins were resolved on SDS-PAGE and visualized by autoradiography. (C) HACul2(WT) was cotranslated in vitro with myc-Rbx1 in the presence of [35S]methionine and incubated with increasing amounts of unlabeled in vitro-translated HA-Cul2ΔC before a neddylation reaction (left panel). HA-Cul2(WT) and myc-Rbx1 were translated in vitro independently in the presence of [35S]methionine. myc-Rbx1 was subsequently incubated with increasing amounts of unlabeled in vitro-translated HA-Cul2ΔC before the addition of radiolabeled in vitro-translated HA-Cul2(WT) and the performance of a neddylation reaction (right panel). All reaction mixtures were immunoprecipitated with anti-HA antibody, resolved on SDSPAGE, and visualized by autoradiography. IP = immunoprecipitation; AR = autoradiography; NEDD8 RxN = neddylation reaction.
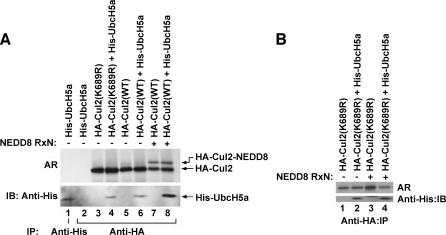
UbcH5a Preferentially Engages Cul2 Modified by NEDD8
Polyubiquitylation of cellular proteins requires the coordinated action of an E3 with E2. The physiological E2 for ECV is yet unknown. However, both Cdc34 and UbcH5a have been shown to effectively support ECV-dependent ubiquitylation of HIFα in vitro, with UbcH5a displaying greater activity [4,10,34]. We asked whether neddylation of Cul2 affected its association with UbcH5a as a potential mechanism of regulating the E3 function of ECV. HA-Cul2(WT) and the neddylation-defective HA-Cul2(K689R) mutant were 35Stranslated in vitro in the presence or in the absence of neddylation conditions and were incubated with purified His-UbcH5a before immunoprecipitation with anti-HA antibody. Bound proteins were resolved on SDS-PAGE and visualized by autoradiography and immunoblotting with anti-His antibody (Figure 3A). Both HA-Cul2(WT) and the HA-Cul2(K689R) mutant coprecipitated equal amounts of His-UbcH5a in the absence of neddylation reactions. Similar results were obtained in vivo from transfected HEK293A cells, in which HA-Cul2(WT) and HA-Cul2(K689R) immunoprecipitated equal amounts of cotransfected FLAG-UbcH5a (data not shown). However, an in vitro neddylation reaction resulted in a dramatic increase of the more slowly migrating neddylated Cul2 and, importantly, of the Cul2-associated UbcH5a. This was not due to the neddylation reaction itself because the amount of His-UbcH5a associating with the neddylation-defective Cul2(K689R) was comparable irrespective of neddylation conditions (Figure 3B). These results suggest that the promotion of Cul2 neddylation increases UbcH5a engagement. It should be mentioned that the detection of endogenous E2s in complex with E3s is notoriously difficult, and we have, to date, been unsuccessful in establishing the interaction of UbcH5a (or Cdc34) with Cul2 (WTor K689R) in the absence of overexpression.
Figure 3.
UbcH5a preferentially binds NEDD8-modified Cul2. (A) 35S-labeled in vitro-translated HA-Cul2(WT) or HA-Cul2(K689R) was mixed with purified His-UbcH5a, subjected to a neddylation reaction, where indicated, and immunoprecipitated with anti-His or anti-HA antibodies. Bound proteins were resolved by SDSPAGE, [35S]Cul2 was visualized by autoradiography, and Cul2-bound UbcH5a was visualized by anti-His immunoblotting. (B) Performed as in (A) with HACul2(K689R) and His-UbcH5a in the absence (-) or in the presence (+) of a neddylation reaction. IP = immunoprecipitation; IB = immunoblot; NEDD8 RxN = neddylation reaction.
HIF1α-Bound ECV Preferentially Contains Neddylated Cul2
An active E3 of the SCF family is invariably associated with its respective E2 and is bound by an F-box protein-specific substrate. A prediction would be that an ECV complex that has recruited HIFα for ubiquitylation would be associated with the neddylated Cul2 that has engaged UbcH5a. To test this hypothesis, HEK293A cells were transiently transfected with plasmids encoding HA-VHL, in combination with HIF1α, and were treated with the proteasome inhibitor MG132 before lysis. Lysates were immunoprecipitated with either anti-HA or anti-HIF1a antibody, resolved by SDSPAGE, and immunoblotted with anti-HIF1a, anti-Cul2, or anti-HA antibodies (Figure 4A). As expected and previously observed [35], VHL coprecipitated both neddylated and unneddylated Cul2, but primarily the latter form (Figure 4A, lane 1). HIF1α coprecipitated VHL as expected but, strikingly and in contrast, preferentially coprecipitated NEDD8-modified Cul2 (Figure 4A, lane 2). In a complementary experiment, HEK293A cells were treated with or without MG132 and immunoprecipitated for endogenous HIF1α (Figure 4B). Proteasomal inhibition, as expected, resulted in the accumulation of HIF1α, which, consistent with overexpression data, preferentially coprecipitated neddylated Cul2 (Figure 4B, lane 2). These results suggest that ECV recognition of HIF1α through VHL is temporally coordinated with the NEDD8 modification of Cul2, resulting in the recruitment of UbcH5a and thereby triggering the ubiquitylation of HIF1α. Notably, we observed a slight but nevertheless significant accumulation of total neddylated Cul2 in the presence of proteasome inhibitor, suggesting that a NEDD8-conjugated form of Cul2 is also unstable and subjected to degradation through the proteasome (Figure 4B, cf. lanes 3 and 4). In support of this notion, neddylated Cul1 and Cul3 were recently shown to be targeted for proteasome-mediated destruction [36,37]. However, this accumulation of neddylated Cul2 in the presence of a proteasomal inhibitor does not account for the Cul2 profile associated with HIF1α (Figure 4B, cf. lane 2 in middle panel and lane 4 in lower panel).
Figure 4.
ECV engaged to HIF1α contains preferentially neddylated Cul2. (A) HEK293A cells were transiently transfected with a mammalian expression plasmid encoding HA-VHL, alone or in combination with a plasmid encoding HIF1α. Cells were treated with the proteasome inhibitor MG132 for 4 hours, lysed, and immunoprecipitated with indicated antibodies. Bound proteins were resolved by SDS-PAGE and immunoblotted with anti-HIF1α (upper panel), anti-Cul2 (middle panel), and anti-HA (lower panel) antibodies. (B) HEK293A cells were treated with MG132 for 4 hours, where indicated, lysed, and immunoprecipitated with anti-HIF1a antibody (lanes 1 and 2). Bound proteins were resolved by SDS-PAGE and immunoblotted with anti-HIF1α (upper panel) and anti-Cul2 (middle panel) antibodies. In parallel, 20 µg of whole-cell extract (lanes 3 and 4) collected before the anti-HIF1α immunoprecipitation experiment was resolved by SDS-PAGE and immunoblotted with anti-Cul2 antibody (lower panel). IP = immunoprecipitation; IB = immunoblot; WCE = whole-cell extract.
HIF1α Binding to ECV Triggers Cul2 Neddylation
We next asked whether the binding of substrate HIFα by the substrate-recruiting protein VHL determined Cul2 neddylation. HEK293A cells were transfected with plasmids encoding the HA full-length VHL (1–213) or the HA-α domain (155–213) of VHL. The latter is incapable of binding HIFα due to the lack of the substrate recognition β domain. Cells were lysed, immunoprecipitated against HA, resolved on SDS-PAGE, and immunoblotted with anti-Cul2 and anti-HA antibodies (Figure 5A). Although the full-length VHL coprecipitated both neddylated and unneddylated Cul2, the a domain of VHL exclusively bound unmodified Cul2 (Figure 5A), suggesting that the VHL engagement of substrate is critical for subsequent neddylation of Cul2. In a complementary experiment, we asked whether an increase in available substrate enhanced the neddylation of Cul2. HEK293A cells were transiently transfected with plasmids encoding HIF1a and T7-VHL and were incubated with the proteasome inhibitor for 4 hours before lysis. Cells were lysed and immunoprecipitated with anti-T7 antibody, and bound proteins were resolved by SDS-PAGE and immunoblotted with anti-HIF1α, anti-Cul2, and anti-T7 antibodies. Although T7-VHL immunoprecipitated mainly unneddylated Cul2, as previously observed, the ectopic expression of HIF1α resulted in a dramatic increase of neddylated Cul2 (Figure 5B). This increase in neddylated Cul2 was dose-dependent on the amount of available HIF1α (Figure 5C). Notably, ectopic expression of HIF1α had negligible effect on the general profile of Cul2, suggesting that ECV is not the sole occupier of Cul2. In support of this notion, Kamura et al. [38] identified several elongin BC/Cul2 complexes that are associated with other potential substrate-recruiting proteins with “loose” F-box motifs. Together, these results demonstrate that HIF1α is not merely recruited to the ECV for ubiquitylation but that it plays an active role in its own demise by triggering the neddylation-dependent activation of the ECV. Furthermore, the ability of VHL to bind HIFα was not affected by the NEDD8 status of Cul2 (data not shown), precluding the possibility that neddylation of Cul2 promotes the ECV engagement of HIFα.
Figure 5.
HIF1α binding to ECV triggers Cul2 neddylation. (A) HEK293A cells were transiently transfected with either empty plasmid (Mock) or plasmids encoding HA-VHL(WT; 1–213) or HA-VHL(155–213). Cells were treated with MG132 for 4 hours, lysed, and immunoprecipitated with anti-HA antibody. Bound proteins were resolved by SDS-PAGE and immunoblotted with indicated antibodies. *Undefined modified VHL. (B) HEK293A cells were transiently transfected with plasmids encoding HIF1α or T7-VHL, or in combination. Cells were treated with MG132 for 4 hours, lysed, and immunoprecipitated with anti-T7 antibody. Bound proteins were resolved by SDS-PAGE and immunoblotted with indicated antibodies. In parallel, 20 µg of whole-cell extract collected before the anti-T7 immunoprecipitation experiment was resolved by SDS-PAGE and immunoblotted with anti-Cul2 antibody (lower panel). (C) HEK293A cells were transiently transfected with mammalian expression plasmids encoding T7-VHL and HIF1α in increasing amounts, as indicated. Cells were treated with MG132 for 4 hours before lysis, lysed, and immunoprecipitated with anti-T7 antibody. Bound proteins were resolved by SDS-PAGE and immunoblotted with indicated antibodies. In parallel, 20 µg of whole-cell extract collected before the anti-T7 immunoprecipitation experiment was resolved by SDS-PAGE and immunoblotted with anti-Cul2 antibody (lower panel). (D) HEK293A cells were transiently transfected with mammalian expression plasmid encoding HA-VHL and maintained for an additional 4 hours under a 21% oxygen environment or switched to a 1% oxygen environment. Cells were lysed and immunoprecipitated with anti-HA antibody. Bound proteins were resolved by SDS-PAGE and immunoblotted with indicated antibodies. IP = immunoprecipitation; IB = immunoblot; WCE = whole-cell extract.
The best determinant of the VHL engagement of HIFα is oxygen tension. Therefore, we utilized hypoxia to abrogate VHL association with HIFα, and then asked whether substrate disengagement from VHL affected Cul2 neddylation. HEK293A cells were transiently transfected with plasmids encoding HA-VHL and maintained under normoxia (21% O2) or hypoxia (1% O2). Cells were lysed, immunoprecipitated with an anti-HA antibody, resolved by SDS-PAGE, and immunoblotted with anti-Cul2 and anti-HA antibodies. Under hypoxia, where VHL fails to recognize HIFα, there was a striking absence of neddylated Cul2, whereas under normoxia, neddylated Cul2 was found in complex with VHL (Figure 5D). This result further confirms our notion that the binding of substrate HIFα by the substrate-recruiting protein VHL determined Cul2 neddylation. It also suggests that the ECV complex is maintained at an energy-conserving “off” position when no substrate is recruited for ubiquitinmediated destruction.
Cullins are the best-characterized substrates of NEDD8/Rub1-conjugating enzymes. Using a baculovirus protein expression system in Sf-9 insect cells, Kamura et al. [40] have shown that conjugation of Rub1 to Cdc53 depended on Rbx1. Similarly, the coexpression of Cul1 or Cul2 with Rbx1 in insect cells resulted in increased neddylation of Cul1/2 [36,39,40]. Morimoto et al. [36] recently showed that Rbx1 binds to the E2 NCE UbcH12, thus providing the mechanism responsible for the Rbx1-mediated neddylation of Cul1 [36]. The promotion of the neddylation process in an in vitro system increased the recruitment of UbcH4 to the SCFβTrCP1 bound to phosphorylated IκBα and overexpressed UbcH4 in HEK293 cells precipitated exclusively by NEDD8-modified Cul1 [19]. Neddylation of Cul1 has also been shown in vitro to dissociate p120CAND1, which, when bound to Cul1, inhibits Skp1 binding, thus preventing SCF complex formation/function. Moreover, Cul1-p120CAND1 and Cul1-Skp1 complexes are mutually exclusive [27–31]. However, other reports have shown that the neddylation-deficient Cul1(K720R) mutant can form an SCF complex and bind IκBα in vivo, arguing against the notion of p120CAND1 inhibiting the association of unneddylated Cul1 to the SCF complex [19,22]. In support, VHL clearly associates with both neddylated and unneddylated (or neddylation-defective K689R) Cul2 (Figures 4 and 5; data not shown), precluding the involvement of p120CAND1 or a p120CAND1-like protein in ECV formation.
VHL recognizes HIFα that has undergone prolyl hydroxylation through a class of PHDs in the presence of oxygen. The recruited HIFα is then subjected to ubiquitylation through the ECV complex, subsequently leading to 26S proteasomemediated destruction. Recent evidence has shown that the NEDD8 modification of Cul2 dramatically increases the E3 activity of ECV [23]. However, the molecular mechanism underlying this process remained unresolved. Here, we propose (Figure 6) that oxygen-dependent HIF1α engagement to ECV is critical in the promotion of Rbx1-mediated neddylation of Cul2. Indeed, ECV complex containing a mutant VHL lacking a substrate-recognition domain is exclusively associated with unneddylated Cul2. Neddylation of Cul2 then preferentially recruits UbcH5a, triggering the ubiquitylation of VHL-bound HIFα. Thus, these events establish a central role for the neddylation of Cul2 in a previously unrecognized, temporally coordinated activation of ECV with the recruitment of its substrate HIFα. How precisely HIFα binding to VHL leads to the neddylation of Cul2 in the context of ECV remains to be an outstanding question yet to be resolved.
Figure 6.
Sequential activation of ECV (see text for details). N8 = NEDD8; NE1 or NAE = NEDD8-activating enzyme; NE2 or NCE = NEDD8-conjugating enzyme; Ub = ubiquitin; UE1 or UAE = ubiquitin-activating enzyme; UE2 or UCE = ubiquitin-conjugating enzyme; UE3 = ubiquitin ligase; eB = elongin B; eC = elongin C; shaded box within HIFα = prolyl-hydroxylated ODD.
Acknowledgements
We thank the members of the Ohh laboratory for their helpful discussions and critical comments.
Footnotes
This work has been supported by the Canadian Cancer Society of the National Cancer Institute of Canada and the Kidney Foundation of Canada. M.O. is a Canada Research Chair in Molecular Oncology.
References
- 1.Kaelin WG., Jr Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2:673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- 2.Ohh M, Kaelin WG., Jr VHL and kidney cancer. Methods Mol Biol. 2003;222:167–183. doi: 10.1385/1-59259-328-3:167. [DOI] [PubMed] [Google Scholar]
- 3.Cockman ME, Masson N, Mole DR, Jaakkola P, Chang GW, Clifford SC, Maher ER, Pugh CW, Ratcliffe PJ, Maxwell PH. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J Biol Chem. 2000;275:25733–25741. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- 4.Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway RC, Conaway JW. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci USA. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, Pavletich N, Chau V, Kaelin WG. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 6.Tanimoto K, Makino Y, Pereira T, Poellinger L. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 2000;19:4298–4309. doi: 10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maynard MA, Ohh M. Von Hippel-Lindau tumor suppressor protein and hypoxia-inducible factor in kidney cancer. Am J Nephrol. 2004;24:1–13. doi: 10.1159/000075346. [DOI] [PubMed] [Google Scholar]
- 8.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 9.Block K, Appikonda S, Lin HR, Bloom J, Pagano M, Yew PR. The acidic tail domain of human Cdc34 is required for p27(Kip1) ubiquitination and complementation of a Cdc34 temperature sensitive yeast strain. Cell Cycle. 2005;4 doi: 10.4161/cc.4.10.2054. [DOI] [PubMed] [Google Scholar]
- 10.Iwai K, Yamanaka K, Kamura T, Minato N, Conaway RC, Conaway JW, Klausner RD, Pause A. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc Natl Acad Sci USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathias N, Johnson SL, Winey M, Adams AE, Goetsch L, Pringle JR, Byers B, Goebl MG. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 13.Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene. 1999;18:6829–6834. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]
- 14.Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 15.Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitinrelated protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osaka F, Saeki M, Katayama S, Aida N, Toh EA, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furukawa M, Zhang Y, McCarville J, Ohta T, Xiong Y. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol Cell Biol. 2000;20:8185–8197. doi: 10.1128/mcb.20.21.8185-8197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, Suzuki H, Shimbara N, Hidaka Y, Osaka F, et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morimoto M, Nishida T, Honda R, Yasuda H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1) Biochem Biophys Res Commun. 2000;270:1093–1096. doi: 10.1006/bbrc.2000.2576. [DOI] [PubMed] [Google Scholar]
- 21.Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci USA. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohh M, Kim WY, Moslehi JJ, Chen Y, Chau V, Read MA, Kaelin WG., Jr An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep. 2002;3:177–182. doi: 10.1093/embo-reports/kvf028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 26.Wu K, Chen A, Pan ZQ. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J Biol Chem. 2000;275:32317–32324. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- 27.Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- 29.Min KW, Hwang JW, Lee JS, Park Y, Tamura TA, Yoon JB. TIP120A associates with cullins and modulates ubiquitin ligase activity. J Biol Chem. 2003;278:15905–15910. doi: 10.1074/jbc.M213070200. [DOI] [PubMed] [Google Scholar]
- 30.Oshikawa K, Matsumoto M, Yada M, Kamura T, Hatakeyama S, Nakayama KI. Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochem Biophys Res Commun. 2003;303:1209–1216. doi: 10.1016/s0006-291x(03)00501-1. [DOI] [PubMed] [Google Scholar]
- 31.Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 32.del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M. AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis Cullin AtCUL1 is required for auxin response. Plant Cell. 2002;14:421–433. doi: 10.1105/tpc.010282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dharmasiri S, Dharmasiri N, Hellmann H, Estelle M. The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J. 2003;22:1762–1770. doi: 10.1093/emboj/cdg190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lonergan KM, Iliopoulos O, Ohh M, Kamura T, Conaway RC, Conaway JW, Kaelin WG., Jr Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morimoto M, Nishida T, Nagayama Y, Yasuda H. Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8-E3 ligase and regulates its stability. Biochem Biophys Res Commun. 2003;301:392–398. doi: 10.1016/s0006-291x(02)03051-6. [DOI] [PubMed] [Google Scholar]
- 37.Wu JT, Lin HC, Hu YC, Chien CT. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol. 2005;7:1014–1020. doi: 10.1038/ncb1301. [DOI] [PubMed] [Google Scholar]
- 38.Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Megumi Y, Miyauchi Y, Sakurai H, Nobeyama H, Lorick K, Nakamura E, Chiba T, Tanaka K, Weissman AM, Kirisako T, et al. Multiple roles of Rbx1 in the VBC-Cul2 ubiquitin ligase complex. Genes Cells. 2005;10:679–691. doi: 10.1111/j.1365-2443.2005.00869.x. [DOI] [PubMed] [Google Scholar]