Abstract
Linkage studies of complex genetic traits raise questions about the effects of genetic heterogeneity and assortative mating on linkage analysis. To further understand these problems, I have simulated and analyzed family data for a complex genetic disease in which disease phenotype is determined by two unlinked disease loci. Two models were studied, a two-locus threshold model and a two-locus heterogeneity model. Information was generated for a marker locus linked to one of the disease-defining loci. Random-mating and assortative-mating samples were generated. Linkage analysis was then carried out by use of standard methods, under the assumptions of a single-locus disease trait and a random-mating population. Results were compared with those from analysis of a single-locus homogeneous trait in samples with the same levels of assortative mating as those considered for the two-locus traits. The results show that (1) introduction of assortative mating does not, in itself, markedly affect the estimate of the recombination fraction; (2) the power of the analysis, reflected in the LOD scores, is somewhat lower with assortative rather than random mating. Loss of power is greater with increasing levels of assortative mating; and (3) for a heterogeneous genetic disease, regardless of mating type, heterogeneity analysis permits more accurate estimate of the recombination fraction but may be of limited use in distinguishing which families belong to each homogeneous subset. These simulations also confirmed earlier observations that linkage to a disease "locus" can be detected even if the disease is incorrectly defined as a single-locus (homogeneous) trait, although the estimated recombination fraction will be significantly greater than the true recombination fraction between the linked disease-defining locus and the marker locus.
Full text
PDF

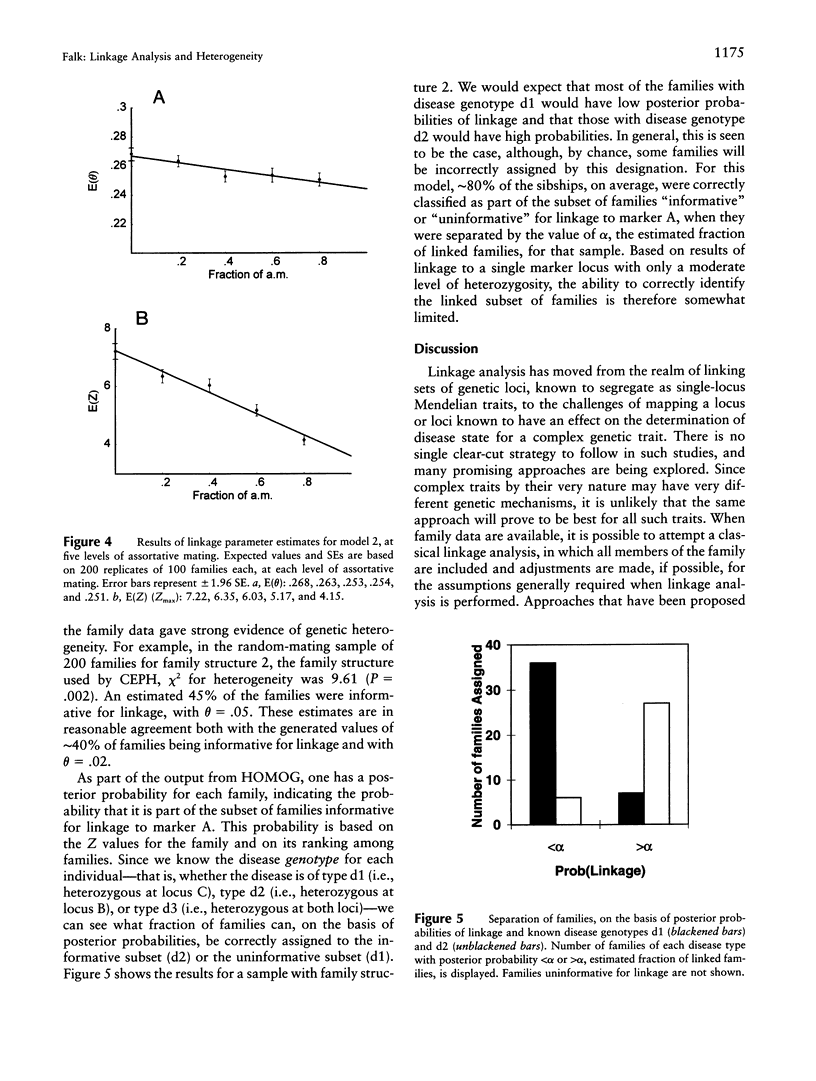



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clerget-Darpoux F., Bonaïti-Pellié C., Hochez J. Effects of misspecifying genetic parameters in lod score analysis. Biometrics. 1986 Jun;42(2):393–399. [PubMed] [Google Scholar]
- Davies J. L., Kawaguchi Y., Bennett S. T., Copeman J. B., Cordell H. J., Pritchard L. E., Reed P. W., Gough S. C., Jenkins S. C., Palmer S. M. A genome-wide search for human type 1 diabetes susceptibility genes. Nature. 1994 Sep 8;371(6493):130–136. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- Durner M., Greenberg D. A., Hodge S. E. Inter- and intrafamilial heterogeneity: effective sampling strategies and comparison of analysis methods. Am J Hum Genet. 1992 Oct;51(4):859–870. [PMC free article] [PubMed] [Google Scholar]
- Egeland J. A., Gerhard D. S., Pauls D. L., Sussex J. N., Kidd K. K., Allen C. R., Hostetter A. M., Housman D. E. Bipolar affective disorders linked to DNA markers on chromosome 11. 1987 Feb 26-Mar 4Nature. 325(6107):783–787. doi: 10.1038/325783a0. [DOI] [PubMed] [Google Scholar]
- Falk C. T. The combined effects of positive assortative mating and selection. Heredity (Edinb) 1971 Aug;27(1):125–136. doi: 10.1038/hdy.1971.77. [DOI] [PubMed] [Google Scholar]
- Goldin L. R. Detection of linkage under heterogeneity: comparison of the two-locus vs. admixture models. Genet Epidemiol. 1992;9(1):61–66. doi: 10.1002/gepi.1370090107. [DOI] [PubMed] [Google Scholar]
- Goldin L. R., Weeks D. E. Two-locus models of disease: comparison of likelihood and nonparametric linkage methods. Am J Hum Genet. 1993 Oct;53(4):908–915. [PMC free article] [PubMed] [Google Scholar]
- Greenberg D. A., Hodge S. E. Linkage analysis under "random" and "genetic" reduced penetrance. Genet Epidemiol. 1989;6(1):259–264. doi: 10.1002/gepi.1370060145. [DOI] [PubMed] [Google Scholar]
- Hodge S. E. Do bilineal pedigrees represent a problem for linkage analysis? Basic principles and simulation results for single-gene diseases with no heterogeneity. Genet Epidemiol. 1992;9(3):191–206. doi: 10.1002/gepi.1370090306. [DOI] [PubMed] [Google Scholar]
- Kelsoe J. R., Ginns E. I., Egeland J. A., Gerhard D. S., Goldstein A. M., Bale S. J., Pauls D. L., Long R. T., Kidd K. K., Conte G. Re-evaluation of the linkage relationship between chromosome 11p loci and the gene for bipolar affective disorder in the Old Order Amish. Nature. 1989 Nov 16;342(6247):238–243. doi: 10.1038/342238a0. [DOI] [PubMed] [Google Scholar]
- Merikangas K. R. Assortative mating for psychiatric disorders and psychological traits. Arch Gen Psychiatry. 1982 Oct;39(10):1173–1180. doi: 10.1001/archpsyc.1982.04290100043007. [DOI] [PubMed] [Google Scholar]
- Ott J. Estimation of the recombination fraction in human pedigrees: efficient computation of the likelihood for human linkage studies. Am J Hum Genet. 1974 Sep;26(5):588–597. [PMC free article] [PubMed] [Google Scholar]
- Risch N., Giuffra L. Model misspecification and multipoint linkage analysis. Hum Hered. 1992;42(1):77–92. doi: 10.1159/000154047. [DOI] [PubMed] [Google Scholar]
- Schork N. J., Boehnke M., Terwilliger J. D., Ott J. Two-trait-locus linkage analysis: a powerful strategy for mapping complex genetic traits. Am J Hum Genet. 1993 Nov;53(5):1127–1136. [PMC free article] [PubMed] [Google Scholar]
- Spence M. A., Bishop D. T., Boehnke M., Elston R. C., Falk C., Hodge S. E., Ott J., Rice J., Merikangas K., Kupfer D. Methodological issues in linkage analyses for psychiatric disorders: secular trends, assortative mating, bilineal pedigrees. Report of the MacArthur Foundation Network I Task Force on Methodological Issues. Hum Hered. 1993 May-Jun;43(3):166–172. doi: 10.1159/000154173. [DOI] [PubMed] [Google Scholar]
- Sribney W. M., Swift M. Power of sib-pair and sib-trio linkage analysis with assortative mating and multiple disease loci. Am J Hum Genet. 1992 Oct;51(4):773–784. [PMC free article] [PubMed] [Google Scholar]
- Vieland V. J., Greenberg D. A., Hodge S. E. Adequacy of single-locus approximations for linkage analyses of oligogenic traits: extension to multigenerational pedigree structures. Hum Hered. 1993 Nov-Dec;43(6):329–336. doi: 10.1159/000154155. [DOI] [PubMed] [Google Scholar]
- Vieland V. J., Hodge S. E., Greenberg D. A. Adequacy of single-locus approximations for linkage analyses of oligogenic traits. Genet Epidemiol. 1992;9(1):45–59. doi: 10.1002/gepi.1370090106. [DOI] [PubMed] [Google Scholar]
- Vieland V., Greenberg D. A., Hodge S. E., Ott J. Linkage analysis of two-locus diseases under single-locus and two-locus analysis models. Cytogenet Cell Genet. 1992;59(2-3):145–146. doi: 10.1159/000133229. [DOI] [PubMed] [Google Scholar]
- Weeks D. E., Lange K. The affected-pedigree-member method of linkage analysis. Am J Hum Genet. 1988 Feb;42(2):315–326. [PMC free article] [PubMed] [Google Scholar]


