Abstract
Botulinum neurotoxins (BoNTs) cause muscle paralysis by selectively cleaving core components of the vesicular fusion machinery within motoneurons. Complex gangliosides initially bind into a pocket that is conserved among the seven BoNTs and tetanus neurotoxin. Productive neurotoxin uptake also requires protein receptors. The interaction site of the protein receptor within the neurotoxin is currently unknown. We report the identification and characterization of the protein receptor binding site of BoNT/B and BoNT/G. Their protein receptors, synaptotagmins I and II, bind to a pocket at the tip of their HCC (C-terminal domain of the C-terminal fragment of the heavy chain) that corresponds to the unique second carbohydrate binding site of tetanus neurotoxin, the sialic acid binding site. Substitution of amino acids in this region impaired binding to synaptotagmins and drastically decreased toxicity at mouse phrenic nerve preparations; CD-spectroscopic analyses evidenced that the secondary structure of the mutated neurotoxins was unaltered. Deactivation of the synaptotagmin binding site by single mutations led to virtually inactive BoNT/B and BoNT/G when assayed at phrenic nerve preparations of complex-ganglioside-deficient mice. Analogously, a BoNT B mutant with deactivated ganglioside and synaptotagmin binding sites lacked appreciable activity at wild-type mouse phrenic nerve preparations. Thus, these data exclude relevant contributions of any cell surface molecule other than one ganglioside and one protein receptor to the entry process of BoNTs, which substantiates the double-receptor concept. The molecular characterization of the synaptotagmin binding site provides the basis for designing a novel class of potent binding inhibitors.
Keywords: synaptotagmin, tetanus
Botulinum neurotoxins (BoNTs) (serotypes A–G) are the causative agents of the disease botulism. The neurotoxins are produced as ≈150-kDa single-chain proteins in Clostridium botulinum and subsequently cleaved by proteases, yielding an ≈100-kDa heavy chain (HC) and an ≈50-kDa light chain (LC). These chains remain connected via a single disulfide bridge, noncovalent interactions, and a HC-derived peptide loop wrapped around the LC. The LCs act as zinc metallopeptidases, which solely hydrolyze one of three SNARE (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor) proteins: vesicle associated membrane protein/synaptobrevin, synaptosomal-associated protein of 25 kDa, or syntaxin. Together, these substrate molecules constitute the core of the vesicular fusion machinery. Thus, cleavage of one of these proteins inhibits the release of neurotransmitters from synaptic vesicles into the synaptic cleft. The HCs mediate the neurospecific binding, uptake by receptor-mediated endocytosis, and transport of the LC across the endosomal membrane into the cytosol, where the LCs encounter their substrates. The ≈50-kDa N-terminal domain of HC (HN) provides the translocation apparatus for the LC (1, 2). The C-terminal half of HC, referred to as the HC fragment, consists of two functionally distinct domains of ≈25 kDa each, HCN and HCC (3). The function of HCN of BoNTs and tetanus neurotoxin (TeNT), a closely related neurotoxin produced by Clostridium tetani, has not yet been elucidated. In contrast, TeNT-HCC has recently been shown to mediate the specific binding of the neurotoxin to nerve terminals at the neuromuscular junction (4). Although BoNTs act locally at motoneurons and thus cause flaccid paralysis, TeNT is transported in a retrograde fashion within the axon and crosses the synaptic cleft into an inhibitory spinal cord neuron to cause spastic paralysis (2).
The process of nerve cell binding and uptake is far less understood compared with the mode of action of the enzymatic domains. Complex gangliosides, a class of glycosphingolipids, which are particularly abundant in the outer leaflet of nerve cell membranes, were recognized to function as receptors for clostridial neurotoxins (CNTs) (5–8). Their important role was recently pinpointed by inhibiting their biosynthesis (9–11). However, the existence of two classes of binding sites distinguished by different affinities and the discovery of protease-sensitive binding to neurons (12, 13) resulted in a double-receptor concept. In a first step complex polysialo gangliosides accumulate CNTs on the plasma membrane surface; and in a second step, protein receptors mediate their endocytosis (14). The identification of synaptotagmins (Syts) I and II as protein receptors for BoNT/B and BoNT/G, and SV2 for BoNT/A supported this concept (15–19). Recent results (20–22) suggested that, unlike BoNTs, TeNT uses glycosyl-phosphatidylinositol (GPI)-anchored glycoproteins of neuronal rafts as receptors.
A cocrystallization approach for BoNT/B and sialyllactose revealed that this trisaccharide occupies a conserved pocket (23), which was shown to be essential for ganglioside interaction of BoNT/A and BoNT/B and TeNT (24, 25). The amino acid sequence and the structure of this site are conserved among all CNTs except BoNT/D and may thus be considered the general ganglioside binding site. However, a second carbohydrate binding site, the sialic acid binding site, was described only for TeNT (26, 27). The sialic acid binding site in TeNT could be used to interact with the sialic acid-containing glycoprotein receptor (21, 24). In contrast, BoNT/B and BoNT/G bind to nonglycosylated Syt-I and Syt-II, and neither BoNT/A nor BoNT/B displays a second carbohydrate binding site (25).
The intention of this study was to identify and characterize the protein receptor binding site of BoNT/B and BoNT/G. We predicted three potential binding sites in BoNT/B by computational analyses and subsequently applied site-directed mutagenesis either to sterically block putative binding sites or to replace amino acids thought to maintain interactions. In this article, we identify the Syt binding site of BoNT/B and BoNT/G at a location similar to the sialic acid binding site in TeNT. Moreover, we show that BoNT/B, the ganglioside and Syt binding sites of which were deactivated, does not exhibit appreciable toxicity. Thus, our data exclude any significant contributions of other cell surface molecules to binding and entry of BoNTs and support the double-receptor concept.
Results
Computer-Based Surface Analysis of the HCC of BoNT/B Reveals Three Potential Syt Binding Sites.
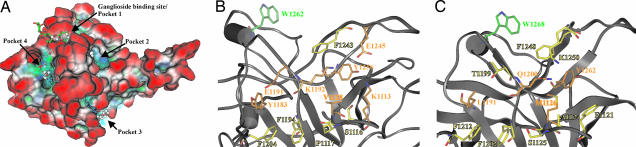
The binding site for Syt-II was recently shown to be located within the HCC of BoNT/B and BoNT/G (15). To precisely identify this interaction site in BoNT/B and BoNT/G, we initially performed a computer-based surface analysis by applying the Molecular Operating Environment (MOE) Site Finder feature (Chemical Computing Group, Montreal, QC, Canada) and SuperStar version 1.5.1 (Cambridge Crystallographic Data Centre, Cambridge, U.K.) that predict hydrophilic and hydrophobic interaction sites and cavities. By means of this procedure, four hollows exhibiting clusters of potential hydrophilic and hydrophobic contact sites were identified, among them the well characterized ganglioside binding pocket (Fig. 1A, pocket 1) (23, 25). Pockets 2 and 4 are located close (≈13 Å) to the ganglioside binding pocket, and pocket 3 is found at the reverse side at a direct distance of 28 Å from the ganglioside binding pocket (Fig. 1A). Pocket 3 was consequently considered to be too far away to mediate the binding to the 20 membrane-proximal residues of Syt, which were determined to be the interacting segment for BoNT/B (16). Pocket 4 covers a surface of ≈790 Å2 and is mainly hydrophobic and encircled by three phenylalanines and two tyrosines with a central lysine (Fig. 1B). Strikingly, pocket 4 matches the position of TeNT's second carbohydrate binding site, the sialic acid binding site (24, 27).
Fig. 1.
Localization of the Syt binding site in BoNT/B and BoNT/G. (A) Computer-based surface analysis of potential Syt binding sites in HCC of BoNT/B. The electrostatic Connolly surface of the HCC of BoNT/B (Protein Data Bank entry 1F31) is displayed. Blue, hydrophilic area; green, hydrophobic area; white, neutral; red, surface exposed. Blue dots represent the recognition of four suitable pockets by the SuperStar algorithm, whereas gray and red balls indicate polar and hydrophobic interaction points found by Site Finder. (B) Residues forming the Syt binding site (pocket 4) of HCC of BoNT/B are shown as sticks. (C) Shown is the homolog Syt binding site of the HCC of BoNT/G. Carbon atoms in orange indicate residues subjected to mutational analysis. The highlighted tryptophan in B and C indicates the location of the ganglioside binding site.
The Analog of the TeNT Sialic Acid Binding Site Mediates the Interaction of BoNT/B and BoNT/G with Syt-I and Syt-II.
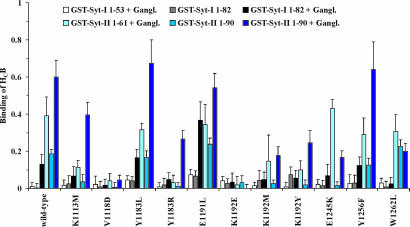
To determine the cavity involved in interaction with the protein receptor, we mutated one central residue in each of the predicted binding sites 2 and 4 of the HC fragment of BoNT/B (HCB). HCB-K1192M, carrying the mutation in pocket 4, exhibited significantly diminished binding to Syt-II, whereas HCB-D1255L, a mutant of pocket 2, did not (Fig. 2 and data not shown). Subsequently, we generated a series of 10 HCB mutants, in which either access to the putative binding pocket 4 was blocked, or the side groups of amino acids predicted to be involved in interactions were isosterically modified to interfere with hydrogen bonding. The binding ability of each of these mutants was studied by GST pull-down experiments. To gain maximum information about the mode of interaction, assays were conducted by using six different bait bead assemblies: the full luminal part of Syt-I (GST-Syt-I 1–53) or Syt-II (GST-Syt-II 1–61) in the presence of gangliosides, and the full luminal part plus transmembrane domain of Syt-I and Syt-II (GST-Syt-I 1–82; GST-Syt-II 1–90) in the absence and presence of gangliosides.
Fig. 2.
GST pull-down assays of BoNT/B HC mutants with Syt-I and Syt-II. The binding ability of HCB mutants (0.06 nmol) was studied by using six different bait bead assemblies (0.75 nmol): the full luminal part of Syt-I (GST-Syt-I 1–53) or Syt-II (GST-Syt-II 1–61) in the presence of gangliosides and the full luminal part plus transmembrane domain of Syt-I and Syt-II (GST-Syt-I 1–82; GST-Syt-II 1–90) in the presence and absence of gangliosides. The amount of bound HCB (molar ratio of GST-Syt to HCB) was analyzed by SDS/PAGE, Coomassie blue staining, and densitometry. Data represent the mean (±SD) of a minimum of three independent experiments.
In agreement with previous findings, wild-type HCB and all of its mutants except HCB-E1191L did not interact with the Syt-I constructs, but did interact with Syt-I 1–82 when assembled together with gangliosides in Triton X-100 micelles (Fig. 2) (15, 16). Syt-II 1–90 showed a lower affinity to HCB in the absence of gangliosides than the transmembrane domain lacking Syt-II 1–61, which binds HCB independent of gangliosides. Micelle incorporation of Syt seemed to sterically hinder HC-fragment interaction with membrane-proximal amino acids. This negative effect turned into the highest affinity observed when Syt-I 1–82 or Syt-II 1–90 and gangliosides were inserted in micelles. Thus, the binding of HCB to both associated components in the micelle membrane is synergistic (Fig. 2). Results obtained with the control HCB-W1262L, an established mutant with a deactivated ganglioside binding site (25), demonstrated that only interactions with Syt-I or Syt-II embedded in mixed Triton X-100/ganglioside micelles were reduced to levels similar to those found when gangliosides were absent. Again, these data argue for two locally separate interactions that are synergistic.
Of the seven residues analyzed in pocket 4 of HCB, the functional groups of Y1256 (hydroxyl group), Y1183 (phenol ring), and E1191 (carboxyl group), all of which occupy positions at the rim of the hollow, did not appear to contribute to Syt binding, because the amounts of bound proteins were not reduced. Furthermore, leucine instead of glutamic acid (E1191L) actually enhanced the interaction with Syt-I. In other words, the native carboxyl group exerted a negative effect on the interaction with Syt-I. Mutation K1113M resulted in moderate effects. In contrast, the import of negative charge by mutations V1118D and K1192E at the central bottom of pocket 4 had a drastic impact on Syt-I or Syt-II interactions independent of membrane assembly or presence of gangliosides. Introduction of positive charge by the replacement of Y1183 by arginine interfered with the binding of HCB to Syt-I or Syt-II in each assembly tested. However, binding of HCB-E1245K to Syt-II lacking its transmembrane domain was not affected, but binding to Syt-II 1–90 was impaired. Presumably, the introduced positive charge of K1245 was repelled by the opposite K1113, which might deteriorate micelle attachment.
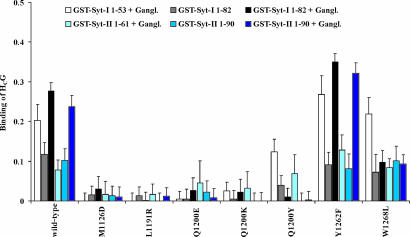
The structure for HC of BoNT serotype G (HCG) is not available yet. To identify the protein binding site in HCG, we initially created a homology model based on the coordinates of the 42% identical HCB (Protein Data Bank entry 1EPW). This model revealed a largely conserved shape for the proposed Syt interaction site, in which F1121, A1124, S1125, M1126, L1191, T1199, Q1200, and K1250 take up the positions of K1113, S1116, P1117, V1118, Y1183, E1191, K1192, and E1245 of HCB, respectively (Fig. 1 B and C). HCB-F1194, F1204, F1243 and Y1256 were found to be conserved (HCG-F1202, F1212, F1248 and Y1262).
In contrast to HCB, HCG displayed a higher affinity for Syt-I 1–53 than Syt-II 1–61 (Figs. 2 and 3) (15). Similar to the interaction of HCB and Syt-II, incorporation of Syt-I or Syt-II into Triton X-100 micelles lacking gangliosides interfered with HCG interaction, but the addition of gangliosides resulted in the highest affinities to Syt-I or Syt-II. When the ganglioside binding pocket of BoNT/G was deactivated by the mutation W1268L, the replacement homologous to W1262L in BoNT/B, the amount of HCG bound to Syt-I 1–82 and Syt-II 1–90 incorporated in ganglioside/Triton X-100 micelles was reduced to levels of Syts embedded in pure Triton X-100 micelles. Again, this finding indicates two independent receptor binding site interactions and argues against a complex formed by the extracellular parts of Syt and ganglioside, as postulated by Kozaki et al. (28). To characterize the homologous pocket of HCG, the amino acids M1126, L1191, Q1200, and Y1262 were replaced analogously to the corresponding HCB residues. The mutations M1126D, L1191R, and Q1200E caused a drastic decrease in affinity to Syt-I and Syt-II, as did their equivalent replacements in HCB with respect to Syt-II binding. Interestingly, replacement of the central residue Q1200 by lysine as found in the corresponding position of HCB (K1192) reduced Syt interaction strongly. The Y1262F mutation had a positive effect on Syt-I and Syt-II binding. The reason for the minor effect of the Q1200Y substitution on binding to either Syt that lacks the transmembrane domain is presently unknown.
Fig. 3.
GST pull-down assays of BoNT/G HC mutants with Syt-I and Syt-II. The binding abilities of HCG mutants (0.06 nmol) were studied by using six different bait bead assemblies (0.75 nmol): the full luminal part of Syt-I (GST-Syt-I 1–53) or Syt-II (GST-Syt-II 1–61) in the presence of gangliosides and the full luminal part plus transmembrane domain of Syt-I and Syt-II (GST-Syt-I 1–82; GST-Syt-II 1–90) in the presence and absence of gangliosides. The amount of bound HCG (molar ratio of GST-Syt to HCG) was analyzed by SDS/PAGE, Coomassie blue staining, and densitometry. Data represent the mean (±SD) of a minimum of three independent experiments.
CD-Spectroscopic Structure Analyses of Mutated HC Fragments.
To exclude the possibility that individual mutations had affected structural elements of the HC fragment, mutants were subjected to far UV CD spectroscopy and thermal-denaturation experiments. CD spectra of all mutants of HCB and HCG were virtually indistinguishable from the respective wild-type spectra [see supporting information (SI) Fig. 6. Secondary structure content was calculated by the algorithm K2d from the CD data between 200 and 240 nm (29). Although HCB and HCG share 43% amino acid identity, the secondary structure content for HCB differs from that of HCG: 21% vs. 31% α-helix, 36% vs. 32% β-sheet, and 42% vs. 37% random coil.
The thermal denaturation of wild-type HCB and HCG and their mutants was irreversible and yielded steep sigmoidal curves (SI Fig. 7). The melting temperatures (Tm), defined by the inflection point, of the HCB mutants lay between 44.6°C and 49.6°C, with wild-type HCB displaying a Tm of 46.8°C. A Tm deviation of ±2.5°C does not indicate structural disturbances evoked by the mutations. All HCG mutants except Q1200Y melted between 50.2°C and 51.6°C. These values were not significantly different from that of wild-type HCG (51.4°C), indicating unaltered secondary structure. This higher stability of HCG vs. HCB goes along with the greater amount of helicity found in HCG. Mutant Q1200Y did not denature until 53.8°C, suggesting an increase in stability that could be interpreted as being due to the new aromatic stacking of the neighboring tyrosines 1200 and 1262.
Pocket 4 Is Essential for the BoNT Activity at the Neuromuscular Junction.
To assess the impact of the various pocket-4 mutations on the biological activity of BoNT, all mutations were incorporated into the full-length BoNT/B or BoNT/G, and activities were determined by the well established mouse phrenic nerve toxicity test. The measured paralytic halftimes were converted to toxicities with respect to the toxicity of the corresponding recombinant wild-type BoNT (Fig. 4).
Fig. 4.
Toxicity of mutated single-chain BoNT/B (scBoNT/B) (black columns) and BoNT/G-Thro (white columns; “Thro” indicates insertion of a thrombin cleavage site) (15) at phrenic-nerve preparations of wild-type mice. To dose–response curves of recombinant wild-type scBoNT/B and wild-type BoNT/G-Thro a power function was fitted. The resulting paralytic halftimes of scBoNT/B or BoNT/G-Thro mutants were converted to the corresponding concentrations of wild-type BoNTs by using the power functions. The toxicities were finally expressed relative to wild-type BoNT.
In BoNT/B, mutations that affected Syt binding in pull-down experiments caused a proportional reduction in activity at the mouse phrenic nerve (Fig. 4). However, BoNT/B-Y1183R was even more active than the wild-type BoNT/B, although the amount of bound HCB-Y1183R was reduced by ≈50%. The reason for this discrepancy might be an improved interaction of the exposed guanidine group with the charged physiological membrane overlying reduced Syt affinity. Interestingly, the V1118D and K1192E mutations caused dramatic drops in activity by factors of ≈116 and ≈312, respectively, which is comparable with mutation of the central W1262 of the ganglioside binding pocket (factor ≈127) (Figs. 4 and 5).
Fig. 5.
Toxicity of scBoNT/B and wild-type BoNT/G-Thro and selected mutants having a deactivated ganglioside binding site (W1262L, W1268L) or Syt binding site (K1192E, Q1200K) at the phrenic nerve preparations of wild-type (black columns) and complex-ganglioside-deficient (white columns) mice. To dose–response curves of recombinant wild-type scBoNT/B and wild-type BoNT/G-Thro a power function was fitted. The resulting paralytic halftimes of scBoNT/B or BoNT/G-Thro mutants were converted to the corresponding concentrations of wild-type BoNTs by using the power functions. The toxicities were finally expressed relative to wild-type BoNT.
In BoNT/G, the majority of the mutations decreased the paralytic activity of the toxin by about factor 4. However, BoNT/G-Y1262F exhibited a slightly increased activity (Fig. 4). These results agree with the changes of Syt binding to HCG mutants in pull-down experiments. The mutant BoNT/G-Q1200K displayed a 20-fold reduction in activity, correlating with its undetectable affinity for micelle-incorporated Syts. This finding was surprising because this mutant incorporated the central lysine mediating a main Syt-II interaction of BoNT/B. In contrast to BoNT/B, none of these mutations reduced the activity to the extent of the substitution W1268L in the ganglioside binding pocket does (77-fold) (Figs. 4 and 5).
Interference of Ganglioside and Syt Interaction Sites of BoNT/B and BoNT/G Nearly Abolishes Toxicity.
The identification of critical residues of the protein receptor binding site allowed for challenging the double-receptor concept of CNTs. If no other sites were involved in neurotoxin binding and uptake into neurons, then a ganglioside and protein receptor binding site double mutant should be inactive. In fact, the effect on the activity of both mutations is synergistic. Because the K1192E and W1262L diminished the activity of BoNT/B 312-fold and 127-fold, respectively, the double mutant consequently exhibited a >50,000-fold reduced activity compared with wild-type BoNT/B (Fig. 5). Analog results were obtained for the double mutant BoNT/G-Thro-Q1200K/W1268L, exhibiting a 2,755-fold reduction at wild-type mice phrenic nerve preparations compared with the single mutations Q1200K and W1268L, which displayed 20-fold and 77-fold decreased activities.
At phrenic nerve preparations from complex ganglioside-deficient mice whose major neuronal ganglioside is GM3, which does not act as a CNT receptor (30), the activity of wild-type BoNT/B and BoNT/G is decreased ≈100-fold (Fig. 5). BoNT/B and BoNT/G having a deactivated ganglioside binding site (W1262L or W1268L) revealed a further 2- to 3-fold reduction in activity, suggesting that the tryptophan to leucine mutations still allowed marginal interaction. In agreement with this interpretation, the BoNT/B-K1192E mutant exhibited only one-third of activity (143,000-fold reduced) at complex ganglioside-deficient phrenic nerve preparations compared with the BoNT/B-K1192E/W1262L mutant at the wild-type phrenic nerves (Fig. 5). Corresponding results, albeit exhibiting less pronounced effects, were obtained for BoNT/G-Q1200K (Fig. 5). Altogether, the interference of both receptor interactions is fatal for the action of BoNTs.
Discussion
A Shallow Pocket Adjacent to the Ganglioside Binding Pocket Mediates the Interaction with Syt.
CNTs bind to complex gangliosides on neuronal cells. The site mediating binding to the carbohydrate portion of gangliosides has been identified for various CNTs (24, 25). It is highly conserved among TeNT and most BoNTs. Molecular details underlying this interaction have also been acquired. In contrast, information about the binding site that ensures adherence of CNTs to their neuronal protein receptor is lacking. In this study, we have addressed this question by using BoNT/B and BoNT/G, which share Syt-I and Syt-II as protein receptors (15). Computer-aided surface analysis and mutational analysis of surface residues demonstrate that a shallow, largely hydrophobic pocket covering ≈790 Å2 in the vicinity of the ganglioside binding pocket at the tip of the HCC mediates the interaction with the protein receptor. This conclusion is based on results that mutations of BoNT/B and BoNT/G residues forming this hollow impaired binding to purified Syt-I and Syt-II and impaired their toxicity at natural target neurons (phrenic nerves).
Recently, Ihara et al. (31) characterized the BoNT/B strain 111, which causes infant botulism. They observed lower binding of HC-BoNT/B 111 than of HC-BoNT/B Okra to GT1b and Syt-II 1–87 embedded in phosphatidylcholine vesicles. The HCC of BoNT/B 111 differs in 20 amino acids from the BoNT/B Okra used in our study. Two residues vary between the Okra and 111 strains in the Syt binding pocket, P1117S and E1191K. The exchange P1117S is less severe and is also found in HCG, but the reversion of polarity by E1191K drastically changes the environment of the pocket. Hence, the reduced binding of HC-BoNT/B 111 compared with HC-BoNT/B Okra can be deduced from these two differences and independently supports our results.
Confirmation for our data was also achieved by the crystal structure of the HCB–Syt-II complex (32). This study revealed that pocket 4 accommodates the C-terminal 10 residues (K51–K60) of Syt-II and that the Syt binding interface of BoNT/B extended into a small neighboring hollow interacting with M46–L50 of Syt-II (32). The luminal domain of Syt-II displays an α-helical conformation, in which the membrane-juxtaposed region of Syt-II is oriented toward the ganglioside binding site. In agreement with our findings, these structural data demonstrate that K1192 (presumably in a salt bridge with E57) and V1118 (hydrophobic interaction with F54) are key players in the interaction with Syt-II.
Conservation of CNT Protein Receptor Binding Sites.
Modeling of the HCC of BoNT/G, using BoNT/B as a template, revealed a largely similar overall shape of the HCCs and the Syt-interaction site. BoNT/B, however, shows a much higher affinity to Syt-II versus Syt-I, whereas BoNT/G interacts with both Syt isoforms similarly but exhibits a slightly weaker affinity compared with the BoNT/B-Syt-II interaction (Figs. 2 and 3) (15). Eight of the 12 residues forming the Syt binding site differ between BoNT/B and BoNT/G. Our results indicate that the change of BoNT/B K1192 to Q1200 in BoNT/G primarily explains the differential affinity of BoNT/B and BoNT/G for Syt-I and Syt-II. Whereas reversion of charge by the K1192E mutation deactivated the Syt binding site in BoNT/B, the substitution of glutamic acid for Q1200 resulted in only a 70% loss of toxicity. But exchange of Q1200 to lysine, as found in BoNT/B, caused the strongest negative effect of all mutations in BoNT/G (95%). Mutational analysis of BoNT/B K1113, E1191 and E1245 indicates that the corresponding residues F1121, T1199, and K1250 in BoNT/G could also contribute to the differential specificity for Syt-I and Syt-II. Furthermore, binding analyses of seven GST-Syt-II 1–61 mutants to wild-type HCB and HCG revealed similar results with one exception. Lysine-57 of GST-Syt-II-E57K being repelled by K1113 and K1192 in HCB did not show a negative effect on binding to HCG (see ref. 32; and T.B., unpublished work). Apparently Syt-II E57 does not contribute to the interaction with BoNT/G. Provided positioning of Syt-I and Syt-II in the binding site is identical, a repulsion of K57 in Syt-II bound to BoNT/G cannot occur because the homolog residues F1121 and Q1200 display no charge. The lack of this ionic interaction may explain the lower affinity of Syt-II to BoNT/G vs. BoNT/B. The dispensable role of the E57 carboxylate, together with the effect of replacing Q1200 by lysine, can also be interpreted by a different positioning of the luminal domain of Syt-I and Syt-II within the Syt binding site in BoNT/G compared with BoNT/B.
Besides the crystal structure of BoNT/B HC, further HC structures have so far been solved only for TeNT and BoNT/A (3, 33). Both display a correspondingly located pocket with shape different from that of the Syt binding site in BoNT/B and BoNT/G, thereby explaining the different protein receptors (15, 16, 18, 19, 21, 24). However, it cannot completely be ruled out that a different site mediates protein receptor binding of the remaining CNTs.
Conclusions on the Binding Mode of CNTs.
Analyses of BoNT/B and BoNT/G having deactivated ganglioside and/or protein receptor binding sites demonstrate that the binding to both ganglioside and Syt receptors is independent but synergistic and is mediated by adjacent but separate binding pockets. In addition, results obtained for BoNT/B and BoNT/G with deactivated protein receptor binding sites in complex-ganglioside-deficient phrenic nerve preparations confirmed this conclusion (Fig. 5). These results rule out (i) that a complex is formed by the extracellular parts of Syt and gangliosides; and (ii) that subsequent to the proposed initial ganglioside binding step, structural changes, as suggested for BoNT/A (34), have to occur to facilitate the interaction with the protein receptor. It is also unlikely that other CNT–glycolipid and CNT–glycoprotein interactions contribute to nerve cell binding. If, as suggested (35), there were such further interactions, at least none of those could compensate for the deactivated ganglioside or protein receptor binding site. Together, the findings of this study are in full agreement with the long-existing double-receptor concept (14).
Another finding in this study is the discovery that the protein receptor binding sites of BoNT/B and BoNT/G are located at the distal tip of the toxin molecule, which ensures that the ganglioside binding pocket faces the membrane. This arrangement allows the neurotoxins to simply dock to the membrane-proximal residues of the Syt luminal domain by lateral diffusion in a ganglioside-bound state without requiring any conformational rearrangement. It is conceivable that then this initial contact triggers the formation of the α-helical conformation of the Syt luminal domain [the isolated luminal domain of Syt-II does not exhibit any structure in solution (32)], and Syt thereby attaches from the C terminus to the N terminus to the protein receptor binding site of the neurotoxin.
The characterization of the Syt receptor binding site allows the development of potent inhibitors restricting access of Syt. Thus, the mechanism of action of the CNTs would be blocked at an early stage. Knowing both ganglioside and protein receptor binding sites, we determined that the crosslinking of potent inhibitors for each site would be synergistic with respect to their inhibitory constant. Furthermore, identification of both receptor sites provides an approach to retarget BoNTs to different cell types by site-directed mutagenesis. Such modified BoNTs could also be used as drug delivery systems (36).
Experimental Procedures
Molecular Modeling.
The homology model of BoNT/G was built up by using the Chemical Computing Group's MOE and by applying the crystal structure of BoNT/B with a resolution of 1.90 Å (Protein Data Bank entry 1EPW) (23) as a template. The target sequence of BoNT/G (HCC, residues 1080–1291) was aligned with BoNT/A (Protein Data Bank entry 3BTA), BoNT/B, and TeNT (Protein Data Bank entry 1FW2) by using the multiple sequence and structural alignment algorithm of MOE with the structural alignment tool enabled and applying the BLOSUM62 substitution matrix. Thus, 10 models were generated based on the BoNT/B structural coordinates. After subjecting each model to coarse energy minimizations and eliminating bad van der Waals contacts between atoms, the best intermediate model was further minimized with the AMBER99 force field until the final energy gradient was found <0.01 kcal/mol·Å. The stereochemical quality of the models was assessed by a Ramachandran plot analysis and structural analysis using the protein report function of the MOE, which searches for disallowed bond angles, bond lengths, and side-chain rotamers.
The binding site analysis of the HCC of BoNT/B and key interaction sites within the cavities was done by using the Site Finder, Contact Statistics, and Multiple Copy Simultaneous Search (MCSS) (37) features of MOE and SuperStar version 1.5.1 (Cambridge Crystallographic Data Centre) (38). All images were created with MOE and Benchware 3D Explorer (Tripos, St. Louis, MO) or Insight 2000 (Accelrys, Cambridge, U.K.).
Plasmid Constructions.
Plasmids encoding the HC fragment (pHCBS and pHCGS) and the full-length BoNT/B and BoNT/G (pBoNTBS and pBoNTGS-Thro) and plasmids encoding the various truncated Syt-I and Syt-II GST fusion proteins used in this study were described in refs. 15 and 25. Mutations in the HC fragments were generated by PCR using suitable primers and pHCBS and pHCGS as template DNA. Correspondingly mutated expression plasmids for full-length BoNTs were generated by swapping DNA fragments between pBoNTBS or pBoNTGS-Thro and mutated pHCBS or pHCGS plasmids, respectively. Nucleotide sequences of all mutants were verified by DNA sequencing.
Purification of Recombinant Proteins.
Wild-type and mutated recombinant HC fragments and full-length BoNTs were produced, the latter under biosafety level 2 containment, by using the Escherichia coli strain M15pREP4 (Qiagen, Hilden, Germany) during 16 h of incubation at room temperature, and were purified on StrepTactin-Sepharose beads (IBA, Göttingen, Germany) according to the manufacturer's instructions. Fractions containing the desired proteins were pooled, frozen in liquid nitrogen, and kept at −70°C. GST fusion proteins were obtained from E. coli TG1 and purified by using glutathione–Sepharose beads (GE Healthcare, Munich, Germany). Fractions containing the desired proteins were pooled and dialyzed against a Tris/NaCl/Triton buffer [20 mM Tris·HCl/150 mM NaCl/0.5% Triton X-100 (pH 7.2)].
GST Pull-Down Assays.
GST fusion proteins (0.075 nmol each) immobilized to 10 μl of glutathione–Sepharose beads at 4°C overnight were incubated with HC fragments or HC fragment mutants (0.06 nmol each) in the absence or presence of a bovine brain ganglioside mixture (18% GM1/55% GD1a/10% GT1b/2% other gangliosides, 5 μg each; Calbiochem, San Diego) in a total volume of 150 μl in Tris/NaCl/Triton buffer for 120 min at 4°C. Beads were collected by centrifugation and washed three times each with 35 bed volumes of the same buffer. Washed pellet fractions and appropriate supernatants were boiled in SDS sample buffer and analyzed together by SDS/PAGE and Coomassie blue staining.
Mouse Phrenic Nerve Toxicity Assay.
The mouse phrenic nerve toxicity assay was set up, as described by Habermann et al. (39) and Wohlfarth et al. (40). Electrical stimulation of the phrenic nerve was continuously performed at a frequency of 1 Hz. Isometric contractions were transformed via a force transducer and recorded with the VitroDat Online software (FMI, Seeheim, Germany). The time required to decrease the amplitude to 50% of the starting value (paralytic halftime) was measured. Recombinant wild-type BoNT/G-Thro was applied in triplicate at final concentrations of 0.6, 2.0, 6.0, 20.0, and 60.0 nM. To the concentration–response curve the power function y = 95.046x−0.2378 (R2 = 0.9722) was fitted. A concentration–response curve with the power function y = 54.368x−0.2947 (R2 = 0.9876) was obtained in the same manner for recombinant single-chain BoNT/B (scBoNT/B) wild type, applying final concentrations of 0.1, 0.3, 1, and 3.2 nM. The resulting paralytic halftimes of scBoNT/B or BoNT/G-Thro mutants were converted to the corresponding concentrations of wild-type BoNTs, by using the equations mentioned above. The toxicities were finally expressed relative to wild-type BoNTs.
CD Spectroscopy.
CD data were collected with a J-810 spectropolarimeter (Jasco, Easton, MD) in a 1-mm pathlength cuvette with a concentration of 5 μM degassed HCB and 2.5 μM degassed HCG in 20 mM sodium phosphate (pH 7.4)/100 mM NaCl. Spectra were recorded at room temperature from 195 to 240 nm with 10 nm/min, sensitivity of 0.01 millidegree, and an accumulation of 3 scans. Temperature-induced denaturation was performed by monitoring the CD signal at 216 nm. HCB and HCG wild-type were measured with temperature increases of 1°C/min from 20°C to 85°C. Because in all cases the denaturation of both wild-type BoNTs happened between 40°C and 60°C, mutants were analyzed between 35°C and 65°C.
Complex-Ganglioside-Deficient Mice.
Mouse phrenic nerves were derived from 20 g NMRI mice (wild-type) and C57BL/6 mice lacking GD3-synthetase (CMP-sialic acid:GM3 α-2,8-sialyltransferase, EC 2.4.99.8) and β-1,4-N-acetylgalactosaminyltransferase (EC 2.4.1.92). Whereas neurons of NMRI mice contain the full set-up of complex polysialo gangliosides, the nerve cells of complex-ganglioside-deficient mice harbor mainly GM3 (30). No difference in paralytic halftime was observed when phrenic nerve preparations of NMRI and wild-type C57BL/6 mice were compared.
Supplementary Material
Acknowledgments
We thank Ulrike Fuhrmann, Kirstin Häfner, Tina Henke, Christina Knorr, Beate Laske, and Jessica Schulze for technical assistance. We thank Rongsheng Jin (Stanford University, Stanford, CA) for his contribution to CD recordings. This work was supported by German Research Council Grant BI 660/2-1 (to T.B.) and by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases Division of Intramural Research (to R.L.P.).
Abbreviations
- BoNT
botulinum neurotoxin
- CNTs
clostridial neurotoxins
- HC
heavy chain
- HC
C-terminal fragment of the HC
- HCC
C-terminal domain of the HC fragment
- HCN
N-terminal domain of the HC fragment
- HCB
HC of BoNT serotype B
- HCG
HC of BoNT serotype G
- LC
light chain
- TeNT
tetanus neurotoxin
- scBoNT/B
single-chain BoNT/B
- SNARE
soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor
- Syt
synaptotagmin.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609713104/DC1.
References
- 1.Bigalke H, Shoer LF. In: Handbook of Experimental Pharmacology. Aktories K, Just I, editors. Vol. 145. Berlin: Springer; 2000. pp. 407–443. [Google Scholar]
- 2.Schiavo G, Matteoli M, Montecucco C. Physiol Rev. 2000;80:717–766. doi: 10.1152/physrev.2000.80.2.717. [DOI] [PubMed] [Google Scholar]
- 3.Umland TC, Wingert LM, Swaminathan S, Furey WF, Schmidt JJ, Sax M. Nat Struct Biol. 1997;4:788–792. doi: 10.1038/nsb1097-788. [DOI] [PubMed] [Google Scholar]
- 4.Herreros J, Lalli G, Schiavo G. Biochem J. 2000;347:199–204. [PMC free article] [PubMed] [Google Scholar]
- 5.van Heyningen WE. Nature. 1974;249:415–417. [Google Scholar]
- 6.Simpson LL, Rapport MM. J Neurochem. 1971;18:1751–1759. doi: 10.1111/j.1471-4159.1971.tb03750.x. [DOI] [PubMed] [Google Scholar]
- 7.van Heyningen WE, Miller PA. J Gen Microbiol. 1961;24:107–119. doi: 10.1099/00221287-24-1-107. [DOI] [PubMed] [Google Scholar]
- 8.Marxen P, Bigalke H. Neurosci Lett. 1989;107:261–266. doi: 10.1016/0304-3940(89)90828-8. [DOI] [PubMed] [Google Scholar]
- 9.Williamson LC, Bateman KE, Clifford JC, Neale EA. J Biol Chem. 1999;274:25173–25180. doi: 10.1074/jbc.274.35.25173. [DOI] [PubMed] [Google Scholar]
- 10.Kitamura M, Takamiya K, Aizawa S, Furukawa K. Biochim Biophys Acta. 1999;1441:1–3. doi: 10.1016/s1388-1981(99)00140-7. [DOI] [PubMed] [Google Scholar]
- 11.Bullens RW, O'Hanlon GM, Wagner E, Molenaar PC, Furukawa K, Plomp JJ, Willison HJ. J Neurosci. 2002;22:6876–6884. doi: 10.1523/JNEUROSCI.22-16-06876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazarovici P, Yavin E. Biochemistry. 1986;25:7047–7054. doi: 10.1021/bi00370a044. [DOI] [PubMed] [Google Scholar]
- 13.Pierce EJ, Davison MD, Parton RG, Habig WH, Critchley DR. Biochem J. 1986;236:845–852. doi: 10.1042/bj2360845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montecucco C. Trends Biochem Sci. 1986;11:314–317. [Google Scholar]
- 15.Rummel A, Karnath T, Henke T, Bigalke H, Binz T. J Biol Chem. 2004;279:30865–30870. doi: 10.1074/jbc.M403945200. [DOI] [PubMed] [Google Scholar]
- 16.Dong M, Richards DA, Goodnough MC, Tepp WH, Johnson EA, Chapman ER. J Cell Biol. 2003;162:1293–1303. doi: 10.1083/jcb.200305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishiki T, Kamata Y, Nemoto Y, Omori A, Ito T, Takahashi M, Kozaki S. J Biol Chem. 1994;269:10498–10503. [PubMed] [Google Scholar]
- 18.Dong M, Yeh F, Tepp WH, Dean C, Johnson EA, Janz R, Chapman ER. Science. 2006;312:592–596. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- 19.Mahrhold S, Rummel A, Bigalke H, Davletov B, Binz T. FEBS Lett. 2006;580:2011–2014. doi: 10.1016/j.febslet.2006.02.074. [DOI] [PubMed] [Google Scholar]
- 20.Herreros J, Lalli G, Montecucco C, Schiavo G. J Neurochem. 2000;74:1941–1950. doi: 10.1046/j.1471-4159.2000.0741941.x. [DOI] [PubMed] [Google Scholar]
- 21.Herreros J, Ng T, Schiavo G. Mol Biol Cell. 2001;12:2947–2960. doi: 10.1091/mbc.12.10.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munro P, Kojima H, Dupont JL, Bossu JL, Poulain B, Boquet P. Biochem Biophys Res Commun. 2001;289:623–629. doi: 10.1006/bbrc.2001.6031. [DOI] [PubMed] [Google Scholar]
- 23.Swaminathan S, Eswaramoorthy S. Nat Struct Biol. 2000;7:693–699. doi: 10.1038/78005. [DOI] [PubMed] [Google Scholar]
- 24.Rummel A, Bade S, Alves J, Bigalke H, Binz T. J Mol Biol. 2003;326:835–847. doi: 10.1016/s0022-2836(02)01403-1. [DOI] [PubMed] [Google Scholar]
- 25.Rummel A, Mahrhold S, Bigalke H, Binz T. Mol Microbiol. 2004;51:631–643. doi: 10.1046/j.1365-2958.2003.03872.x. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Fotinou C, Black I, Fairweather NF, Charles IG, Watts C, Hewitt E, Isaacs NW. J Biol Chem. 2000;275:8889–8894. doi: 10.1074/jbc.275.12.8889. [DOI] [PubMed] [Google Scholar]
- 27.Fotinou C, Emsley P, Black I, Ando H, Ishida H, Kiso M, Sinha KA, Fairweather NF, Isaacs NW. J Biol Chem. 2001;276:32274–32281. doi: 10.1074/jbc.M103285200. [DOI] [PubMed] [Google Scholar]
- 28.Kozaki S, Kamata Y, Watarai S, Nishiki T, Mochida S. Microb Pathog. 1998;25:91–99. doi: 10.1006/mpat.1998.0214. [DOI] [PubMed] [Google Scholar]
- 29.Andrade MA, Chacon P, Merelo JJ, Moran F. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 30.Kawai H, Allende ML, Wada R, Kono M, Sango K, Deng C, Miyakawa T, Crawley JN, Werth N, Bierfreund U, Sandhoff K, Proia RL. J Biol Chem. 2001;276:6885–6888. doi: 10.1074/jbc.C000847200. [DOI] [PubMed] [Google Scholar]
- 31.Ihara H, Kohda T, Morimoto F, Tsukamoto K, Karasawa T, Nakamura S, Mukamoto M, Kozaki S. Biochim Biophys Acta. 2003;1625:19–26. doi: 10.1016/s0167-4781(02)00537-7. [DOI] [PubMed] [Google Scholar]
- 32.Jin R, Rummel A, Binz T, Brunger AT. Nature. 2006;444 doi: 10.1038/nature05387. in press. [DOI] [PubMed] [Google Scholar]
- 33.Lacy DB, Tepp W, Cohen AC, DasGupta BR, Stevens RC. Nat Struct Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- 34.Yowler BC, Schengrund CL. Biochemistry. 2004;43:9725–9731. doi: 10.1021/bi0494673. [DOI] [PubMed] [Google Scholar]
- 35.Montecucco C, Rossetto O, Schiavo G. Trends Microbiol. 2004;12:442–446. doi: 10.1016/j.tim.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Bade S, Rummel A, Reisinger C, Karnath T, Ahnet-Hilger G, Bigalke H, Binz T. J Neurochem. 2004;97:1465–1492. doi: 10.1111/j.1471-4159.2004.02844.x. [DOI] [PubMed] [Google Scholar]
- 37.Miranker A, Karplus M. Proteins. 1999;11:29–34. doi: 10.1002/prot.340110104. [DOI] [PubMed] [Google Scholar]
- 38.Verdonk ML, Cole JC, Taylor R. J Mol Biol. 1999;289:1093–1108. doi: 10.1006/jmbi.1999.2809. [DOI] [PubMed] [Google Scholar]
- 39.Habermann E, Dreyer F, Bigalke H. Naunyn-Schmiedebergs Arch Pharmacol. 1980;311:33–40. doi: 10.1007/BF00500299. [DOI] [PubMed] [Google Scholar]
- 40.Wohlfarth K, Goschel H, Frevert J, Dengler R, Bigalke H. Naunyn-Schmiedebergs Arch Pharmacol. 1997;355:335–340. doi: 10.1007/pl00004951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.