Abstract
The ability to isolate prostate stem cells is essential to explore their role in prostate development and disease. In vitro prostate colony- and sphere-forming assays were used to quantitatively measure murine prostate stem/progenitor cell enrichment and self-renewal. Cell surface markers were screened for their ability to positively or negatively enrich for cells with enhanced growth potential in these assays. Immunohistochemical and FACS analyses demonstrate that specific cell surface markers can be used to discriminate prostate stromal (CD34+), luminal epithelial (CD24+CD49f−), basal epithelial (CD24+CD49f+), hematopoietic (CD45+, Ter119+), and endothelial (CD31+) lineages. Sorting for cells with a CD45−CD31−Ter119−Sca-1+CD49f+ antigenic profile results in a 60-fold enrichment for colony- and sphere-forming cells. These cells can self-renew and expand to form spheres for many generations and can differentiate to produce prostatic tubule structures containing both basal and luminal cells in vivo. These cells also localize to the basal cell layer within the region of the gland that is proximal to the urethra, which has been identified as the prostate stem cell niche. Prostate stem cells can be isolated to a purity of up to 1 in 35 by using this antigenic profile. The remarkable similarity in cell surface profile between prostate and mammary gland stem cells suggests these markers may be conserved among epithelial stem cell populations.
Keywords: CD49f, integrin α6, Sca-1, CD24 heat-stable antigen, stem cell niche
Stem cells are of interest clinically because of their potential to repair damaged tissues, treat degenerative diseases, and because of their purported role in tumor initiation. The ability to identify and isolate stem cells is necessary to study their specialized biology. Enrichment for many types of tissue stem cells has been achieved by using cell surface markers. Murine hematopoietic stem cells can be enriched by sorting Lin−Thy-1loSca-1+ckit+ cells from the bone marrow (1). Recent studies suggest that even better purity can be achieved by further sorting based on expression of the SLAM family receptors CD150 and CD48 (2). Bronchioavelolar stem cells can be isolated from their niche at the bronchioalveolar duct junction (BADJ) by sorting cells with a CD45−CD31−Sca-1+CD34+ profile (3). Data from two recent reports show that mouse mammary stem cells possess a Lin−Sca-1+CD140a−CD24+CD49f+CD29+ cell surface profile and can be isolated to a purity of up to 1 in 20 by using subsets of these markers (4, 5).
The presence of stem cells in the prostate first was proposed to explain the seemingly inexhaustible capacity of the organ to regenerate during androgen cycling experiments (6). The identification of side-population cells and replication quiescent BrdU label-retaining cells further suggests that stem cells exist in the gland (7, 8). Several studies have enriched for primitive prostate cells by using cell surface markers. Richardson et al. (9) demonstrated that the CD44+/α2β1hi/CD133+ human prostate cell subpopulation is 10-fold enriched over CD44+/α2β1hi/CD133− cells for colony-forming activity in vitro. Sca-1 also has been used to enrich for murine prostate cells with enhanced prostate tubule forming capacity in vivo (10, 11). Sca-1+ cells possess several properties of stem cells, including replication quiescence, multilineage differentiation capacity, and localization in the region of prostatic ducts proximal to the urethra that has been identified as the prostate stem cell niche. Sca-1 also is expressed in fetal prostate epithelium, demonstrating it is expressed by prostate stem cells from very early stages of prostate development (12). Here, we demonstrate that the CD45−CD31−Ter119−Sca-1+CD49f+ prostate cell subpopulation is enriched for cells capable of both colony and sphere formation in vitro. These cells can self-renew to form spheres for multiple generations and can differentiate to produce prostatic tubule structures containing both basal and luminal cells in vivo.
Results
Identification of Prostate Cell Lineage Markers.
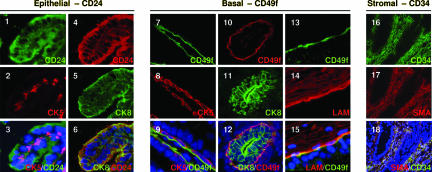
Each of the murine prostate basal, luminal, and stromal cell types can be identified definitively in situ by using antibodies against intracellular proteins, but cell surface markers to identify and isolate each cell type by FACS have not been identified. A panel of antibodies against cluster designation (CD) antigens were screened to identify cell surface markers that can discriminate each of the prostate cell lineages. Immunohistochemical analysis for CD24 (heat stable antigen) demonstrates that it colocalizes with both prostate basal [cytokeratin (CK) 5] and luminal (CK8) cell markers and, therefore, can be used as a pan-epithelial cell marker (Fig. 11–6). CD24 also is expressed by both basal and luminal epithelial cells of the human prostate and mouse mammary gland (13, 14). CD49f (integrin α6) appears to specifically stain the basal surface of CK5+ basal cells (Fig. 1 7–9). This polarization of CD49f expression has been reported for basal cells in other tissues and is elucidated further here by its colocalization with its extracellular matrix ligand, laminin (Fig. 1 13–15) (13, 15, 16). CD49f does not costain with the luminal cell marker CK8 (Fig. 1 10–12) and therefore can be used as a marker for basal epithelial cells. Lastly, colocalization of CD34 with the prostate stromal cell marker smooth muscle actin (SMA) (Fig. 1 16–18) demonstrates it can be used to identify stromal cells.
Fig. 1.
Identification of prostate cell lineage markers. Frozen prostate tissue sections from 10-week-old mice were stained with antibodies against cell surface markers (CD24, CD49f, and CD34) vs. standard intracellular lineage markers (CK5, CK8, and SMA). (Original magnification: ×200.)
FACS analysis for each marker was performed to quantify the percentage of each population in the prostate and investigate whether there is any overlap in expression between each marker. Supporting information (SI) Fig. 6A demonstrates that CD24, CD49f, and CD34 are expressed by 59.0 ± 4.5%, 2.6 ± 0.7%, and 8.0 ± 2.2% of prostate cells, respectively. Nonprostate cell lineages within the prostate also can be identified by FACS with antibodies against CD45 (hematopoietic), Ter119 (red blood cell), and CD31 (endothelial). Pairwise analysis for all markers shows that the stromal cell marker CD34 does not significantly overlap with either epithelial marker CD24 or CD49f. Nearly all CD49f+ cells express CD24 as predicted based on in situ analysis. Small populations of CD24+ and CD49f+ cells also express the nonprostate cell lineage markers (CD45, Ter119, and CD31, collectively called “Lin”), which is consistent with studies showing hematopoietic and endothelial cells express CD24 and CD49f (17, 18). These data suggest that it should be possible to isolate basal and luminal cells by sorting the CD24+CD49f+ and CD24+CD49f− fractions of the prostate, respectively. CK5 and CK8 staining of cytospins prepared from each fraction demonstrates that >70% of CD24+CD49f+ cells express CK5 (SI Fig. 6B Left). Conversely, the majority of CD24+CD49f− cells are CK8+ (SI Fig. 6B Right), and <4% of these cells express CK5 (data not shown).
Lin−Sca-1+CD49f+ (LSC) Cells Are >60-Fold Enriched for Colony-Forming Activity in Vitro.
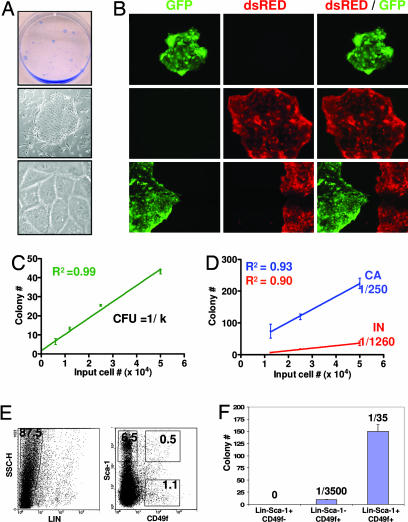
Human prostate epithelial cells can be grown as clonal colonies in vitro most effectively when cocultured with irradiated fibroblast feeder layers in serum-free medium (19, 20). These growth conditions therefore were used to quantitatively screen a series of cell surface markers for their ability to positively or negatively enrich for colony-forming cells. When dissociated prostate cells were plated on top of irradiated 3T3 feeder cells, colonies containing cells with tight epithelium-like borders that express epithelial-specific cytokeratins were observed (Fig. 2A and SI Fig. 7A). To determine whether prostate epithelial colonies are of clonal origin, mixtures of prostate cells from transgenic animals expressing GFP or dsRED under regulation of the β-actin promoter were cocultured in several wells of a six-well plate in two separate experiments. Only 8 of the 188 (4%) colonies observed 10 days later contained both GFP+ and dsRED+ cells, indicating colonies are predominantly of clonal origin in this assay (Fig. 2B). When serial dilutions of wild-type prostate cells from C57BL/6 animals were plated, the number of colonies grown at each dilution was proportional to the number of input cells (Fig. 2C). This linear distribution is further indicative of a clonal outgrowth model and suggests that this assay can be used quantitatively. The number of prostate cells within a population capable of colony outgrowth, which we define here as the colony-forming unit (cfu), can be calculated by taking the inverse of the slope (k) of the best-fit line (cfu = 1/k).
Fig. 2.
LSC cells are enriched for colony-forming activity in vitro. (A Top) Overview of prostate colonies stained with trypan blue. (A Middle and Lower) Light microscopy images of colonies. (Original magnification: ×100 and ×1,000.) (B) Prostate cells were dissociated from four β-actin GFP and four β-actin dsRED transgenic mice in two independent experiments. (B Top and Middle) Representative images of colonies grown in wells where 2 × 104 dsRED+ or GFP+ cells were plated separately. (B Bottom) Images of colonies observed when 1 × 104 dsRED+ and 1 × 104 GFP+ cells were plated together. (Original magnification: ×100.) (C) Prostate cells from four animals were plated in vitro in a dilution series ranging from 0.3 to 5 × 104 cells per well. Graph shows the number of colonies grown at each dilution plotted versus the input cell number. (D) Four of eight 10-week-old male animals were surgically castrated. Prostate cells were harvested from castrated (CA) and intact (IN) animals two weeks later and plated in the dilution series indicated. Graph shows the number of colonies grown plotted versus the input cell number in two pooled experiments. (E) FACS plots show gates drawn for sorting of LSC, Lin−Sca-1+CD49f−, and Lin−Sca-1−CD49f+ subpopulations. (F) Prostate cells from 10 animals were sorted into the populations shown in E. Each population was plated in several replicates at a density of 5,000 cells per well. Bar graph shows the number of colonies that grew out 10 days later.
Castration has been shown to induce apoptosis in mature, androgen-dependent prostate cells and concomitantly enrich for stem and progenitor cells (21). To determine whether such enrichment can be measured using this assay, serial dilutions of cells from intact or castrated animals were compared for colony-forming activity in vitro. Pooled data from two independent experiments demonstrates that cells from castrated animals are >5-fold enriched for colony-forming activity when compared with cells from intact animals (Fig. 2D). This suggests that this assay is sensitive to measuring differences in the number of input stem/progenitor cells.
We and others have reported that Sca-1 can be used to enrich for prostate cells with enhanced growth activity in an in vivo prostate regeneration assay (10, 11). Pooled data from two experiments shows that sorting for Sca-1+ cells yields a cfu of 1:1,000, which represents a 2-fold enrichment over unfractionated prostate cells that were subjected to similar sorting conditions (cfuunfractionated = 1:2,200, SI Fig. 7B). Sca-1− cells have negligible colony-forming activity. When basal, luminal, and stromal cell fractions were sorted based on the profiles identified in SI Fig. 6, all of the colony-forming activity of the prostate was contained within the CD24+CD49f+ basal cell fraction (SI Fig. 7C). CD24+CD49f− luminal cells and CD34+ stromal cells have negligible colony-forming activity (SI Fig. 7 C and D). Sorting for CD24+CD49f+ cells results in a cfu of 1:400 and a 6-fold enrichment for colony-forming activity. These data suggest that prostate colony-forming cells possess a Sca-1+CD24+CD49f+CD34− antigenic profile.
FACS analysis for Sca-1, CD24, CD49 and CD34 reveals that all Sca-1+CD49f+ prostate cells are CD24+ and CD34− (data not shown). CD24 and CD34 therefore were omitted in future studies because they do not subfractionate the Sca-1+ CD49f+ population. Sorting for Sca-1+CD49f+ cells results in an average cfu of 1:140 in three independent experiments, which corresponds to a 15-fold enrichment over unfractionated cells (SI Fig. 7E). Because nonprostate cell lineages within the prostate also express CD49f and Sca-1 (SI Fig. 6A), we projected that removal of these cells from the Sca-1+CD49f+ fraction would result in further enrichment for primitive prostate cells. Prostate cells therefore were sorted into LSC, Lin−Sca-1+CD49f−, and Lin−Sca-1−CD49f+ populations (Fig. 2E) and plated in several replicates at a single density of 5,000 cells per well because only small cell numbers were recovered in these experiments. Fig. 2F shows that nearly all of the colony-forming cells of the prostate are contained within the LSC population, which represents only 0.5% of total prostate cells. The LSC population yielded an enrichment of >60-fold for colony-forming cells, where an average of 1 in 35 cells were capable of colony formation in three independent experiments. Prostate colony-forming cells therefore can be highly enriched by sorting cells with a LSC profile (summarized in SI Fig. 7F).
LSC Cells Localize to the Basal Cell Layer Within the Proximal Region of the Murine Prostate.
The preferential survival of basal cells after androgen depletion and prostate involution has led to the hypothesis that prostate stem cells reside in the basal cell layer (21). BrdU pulse–chase experiments showing that the ductal regions of the prostate proximal to the urethra are enriched for slow-cycling label-retaining cells suggest an alternative model where prostate stem cells may exhibit a regional rather than basal-specific localization (8).
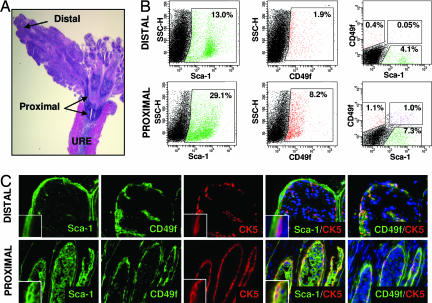
Immunohistochemical and FACS analyses were performed to determine the localization of LSC cells in the prostate. Fig. 3B shows that the percentage of Sca-1+ and CD49f+ populations are both higher in the proximal than distal region as reported (2-fold and 4-fold higher, respectively) in refs. 10, 11, and 22. The LSC population demonstrates an even more dramatic enrichment. The percentage of LSC cells is >20-fold higher in the proximal than distal region (Fig. 3B). Immunohistochemical staining to determine the lineage status of this population demonstrates that CD49f+Sca-1+ cells in the proximal region express the basal cell marker CK5 (Fig. 3C Lower). Distal CD49f+ cells express CK5 but not Sca-1. Sca-1 is expressed only by stromal cells in this region (Fig. 3C Upper). This corresponds with FACS data showing there are small populations of cells positive for CD49f (0.4%) or Sca-1 (4.1%), but negligible if any double-positive cells in the distal region (Fig. 3B). These data suggest that LSC cells must localize to the basal cell layer specifically within the proximal region of the prostate, integrating both prevailing hypotheses for the localization of prostate stem cells. Furthermore, because the CD49f+Sca-1− basal cells in the distal regions of the gland do not appear to have significant colony-forming activity in vitro (Fig. 2F), these data also suggest that the basal cell layer contains functionally distinct subpopulations of cells and that these functional differences may be dictated by regional localization.
Fig. 3.
LSC cells localize to the basal cell layer within the proximal region of the prostate. (A) H&E stain of a longitudinal section from a 10-week-old mouse prostate. Arrows denote regions of the anterior prostate proximal and distal to the urethra (URE). (Original magnification: ×10.) (B) Tissue from the proximal and distal regions of four mouse prostates was microdissected and digested to make dissociated cell suspensions. (B Left and Center) FACS analysis was performed to compare Sca-1 (Left) and CD49f (Center) expression in each region. (B Right) Gated on Lin− cells and indicate the percentage of LSC, Lin−Sca-1+CD49f− and Lin−Sca-1−CD49f+ populations in each region of the prostate. (C) Fluorescence microscopy images show Sca-1 (green), CD49f (green), CK5 (red), and DAPI nuclear counterstain (blue) in the proximal and distal regions of the prostate. (C Insets) Magnified views of cells within each image. (Original magnification: ×200.)
The LSC Subpopulation Is Enriched for Sphere-Forming Cells with Extensive Self-Renewal Capacity in Vitro.
Assays developed to measure neural and mammary stem cell self-renewal in vitro propagate these cells as spheres in an attempt to preserve their specialized biology (23, 24). Several studies have shown that human prostate cells can form epithelial spheroids when cocultured with stromal cells and suspended in Matrigel in vitro (25–27). This strategy was adapted to develop a method for measuring murine prostate stem cell self-renewal.
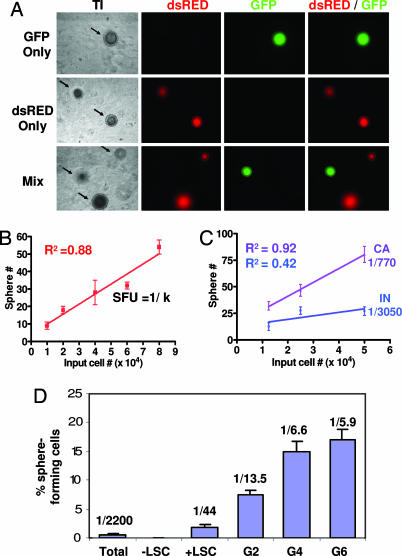
When dissociated murine prostate cells were mixed with fetal stromal cells (urogenital sinus mesenchyme, UGSM) and suspended in Matrigel and PrEGM as described in Materials and Methods, sphere-like structures as shown in Fig. 4A Left were generated. Detailed characterization of this assay will be published separately (unpublished data). To determine whether prostate spheres are of clonal origin, GFP+ and dsRED+ prostate cells were cocultured as in the colony assay. Of 469 spheres counted, no spheres containing both dsRED+ and GFP+ cells were observed (Fig. 4A). Serial dilution experiments further illustrate sphere clonality because there is a linear distribution between the number of input cells and the number of spheres observed (Fig. 4B). Matrigel culture may favor clonal outgrowth because spheres are immobilized and cannot merge together as was recently reported in the neurosphere assay (28). The sphere-forming capacity of cells from castrated and intact animals also was compared to determine whether enrichment for primitive cells results in an enrichment for sphere-forming activity. Fig. 4C shows that castration results in a 4-fold enrichment for sphere-forming cells, suggesting that this assay is also useful for measuring primitive cell enrichment.
Fig. 4.
The LSC subpopulation is enriched for sphere-forming cells that can self-renew in vitro. (A Top and Middle) Prostate cells (8 × 104) from β-actin GFP (Top) or β-actin dsRED (Middle) transgenic mice were mixed with UGSM cells, suspended in matrigel, and plated in vitro. (A Bottom) GFP+ and dsRED+ cells (4 × 104 for each) also were mixed together and added to UGSM in matrigel to investigate whether spheres are of clonal origin. (Original magnification: ×100.) (B) Prostate cells from four mice were mixed with UGSM, suspended in matrigel, and plated in a dilution series ranging from 1 × 104 to 8 × 104 cells. Graph shows the number of spheres grown at each dilution plotted versus the input cell number. (C) Four of eight 10-week-old animals were castrated surgically. Prostate cells were harvested from castrated (CA) and intact (IN) animals two weeks later and mixed with UGSM. Mixtures were suspended in matrigel and plated in the dilution series indicated. Graph shows the number of spheres grown at each dilution plotted versus the input cell number. (D) LSC cells (+LSC) and the fraction depleted of LSC cells (−LSC) were sorted from 10 animals in two separate experiments. Two replicates of 3,500 cells from each population were mixed with UGSM cells, suspended in matrigel, and plated in vitro. Spheres generated from LSC cells were passaged for six generations. Bar graph shows the percentage of sphere-forming cells and the sphere-forming units observed for each primary cell subpopulation (Total, +LSC, and −LSC) as well as for three representative subsequent generations when LSC cells were passaged (G2, G4, and G6).
To determine the sphere-forming potential of the LSC prostate cell subpopulation, these cells were sorted and suspended in Matrigel with UGSM. An average sphere-forming units (sfu) of 1 in 44 was observed in two independent experiments, which represents a 55-fold enrichment over unfractionated prostate cells subjected to similar sorting conditions (sfuunfractionated = 1:2,200; Fig. 4D). Only rare spheres were observed when the fraction depleted of LSC cells were plated in this assay. These data are highly consistent with results attained by using the in vitro colony assay. To assess the self-renewal potential of LSC cells, cells were dissociated from spheres and replated in Matrigel. Fig. 4D shows that these cells were capable of forming spheres for at least six generations in this experiment, where the sphere-forming efficiency ranged from 7–20%. Independent experiments have shown that spheres can be passaged for >10 generations. This demonstrates that the LSC cell subpopulation possesses extensive capacity for self-renewal and expansion in this assay.
LSC Cells Can Differentiate to Produce Prostatic Tubule Structures Containing Both Basal and Luminal Cells in Vivo.
Using a tissue recombination procedure, Cunha and Lung (29) first demonstrated that prostate tissue can be grown de novo when fragments of adult rodent prostate tissue are combined with fragments of UGSM and implanted under the kidney capsule of immunodeficient mice. We have since modified this system to use dissociated cells rather than tissue fragments (30), and others have shown that prostate ducts will regenerate when using a shorter incubation period and a simpler method of injecting cells s.c. in Matrigel (31, 32).
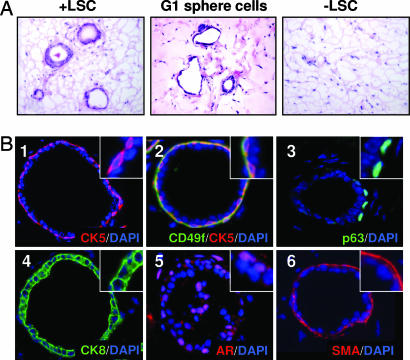
Primary and cultured LSC cells were evaluated for their capacity to form prostate ductal structures by using this in vivo prostate regeneration method. LSC cells and the fraction depleted of these cells were sorted from β-actin dsRED and β-actin GFP animals as in the colony and sphere assays. Three thousand dsRED+ LSC cells were mixed with 3,000 GFP+ LSC cells, and 3,000 dsRED+ LSC-depleted cells were mixed with 3,000 GFP+ LSC-depleted cells. Each sample was mixed with UGSM and injected in vivo. Sphere cells from the first generation of the experiment shown in Fig. 4D also were mixed with UGSM and injected in vivo. Fig. 5A shows that primary LSC cells as well as the sphere cells they produced in vitro are capable of regenerating prostatic tubule structures in vivo. No tubules were observed in grafts grown from the fraction depleted of LSC cells. This suggests that all of the in vivo regenerative activity of the prostate is contained within the LSC subpopulation and that this population can be expanded in vitro without loss of regenerative activity.
Fig. 5.
LSC cells can differentiate to produce prostatic tubules containing both basal and luminal cells in vivo. (A) LSC cells (+LSC) and the fraction depleted of these cells (−LSC) were sorted from 10 β-actin dsRED and 10 β-actin GFP animals. Three thousand dsRED+ LSC cells were mixed with 3,000 GFP+ LSC cells, and 3,000 dsRED+ LSC-depleted cells were mixed with 3,000 GFP+ LSC-depleted cells. Three replicates of each sample were mixed with 2 × 105 UGSM cells and injected in vivo. Fifty thousand sphere cells harvested from the first passage of the experiment shown in Fig. 4D also were mixed with 2 × 105 UGSM and injected in vivo. Images show H&E stains of sections cut from each graft 5 weeks later. (Original magnification: ×200.) (B) Tissue sections of grafts regenerated from primary and cultured LSC cells were stained with antibodies against CK5 (red, 1), CD49f (green, 2), p63 (green, 3), CK8 (green, 4), AR (red, 5), and SMA (red, 6). Each tissue section was counterstained with DAPI (blue) nuclear stain. (Original magnification: ×200.)
Experiments in which mixtures of differentially marked cells were implanted in the in vivo prostate regeneration system have demonstrated that regenerated tubules are of clonal origin because chimeric tubules are observed rarely (11, 12, 32). Likewise, no chimeric tubules containing both dsRED+ and GFP+ cells were observed in these experiments (SI Fig. 8). Fig. 5B shows that tubules regenerated from both primary and cultured LSC cells possess normal lineage marker expression patterns. Regenerated tubules contain populations of basal cells that express CK5 (Fig. 5B1), CD49f (Fig. 5B2) and p63 (Fig. 5B3). Each tubule also contains CK8+ luminal cells (Fig. 5B4) as well as cells that express low levels of the androgen receptor (Fig. 5B5). A layer of SMA+ stromal cells also surrounds each ductal structure (Fig. 5B6). Because regenerated tubules are clonal, the presence of both basal and luminal cells in each tubule indicates that LSC cells are capable of multilineage differentiation, which is a defining property of stem cells.
Discussion
Stem cells are defined by their unique capacity for self-renewal and multilineage differentiation. We find that CD45−CD31−Ter119−Sca-1+CD49f+ prostate cells can self-renew to form spheres for many generations and can differentiate to produce prostatic tubule structures containing both basal and luminal cells in vivo. These cells also localize to the putative prostate stem cell niche in the proximal region of the gland. We therefore conclude that prostate stem cells can be highly purified by using this antigenic profile.
An interesting implication of this study is the remarkable conservation of antigenic profile between prostate stem cells and other types of tissue stem cells. Sca-1 is present on hematopoietic, lung, and mammary stem cells (1, 3, 33). CD49f/integrin α6 shows an even wider distribution amongst stem cell populations. Enrichment for skin stem cells can be achieved by sorting cells that express high levels of CD49f or its subunit pair CD29/integrin β1 (34, 35). A recent study by Stingl et al. (4) showed that mammary stem cells express a CD45−Ter119−CD31−CD140a−CD24medSca-1loCD49fhi cell surface phenotype, which is very similar to the CD45−Ter119−CD31−CD34−CD24+Sca-1+CD49f+ profile identified here for the prostate stem cell, because CD34 is the stromal marker counterpart for CD140a in the prostate. Microarray experiments performed by several independent groups also have shown CD49f is overexpressed consistently in hematopoietic, neural, and embryonic stem cells (36).
The conservation of these markers on stem cells suggests they also may function in maintaining the stem cell phenotype. Integrin α6 pairs with either β1 or β4 to form an integrin receptor that recognizes the basement membrane and extracellular matrix (ECM) protein laminin as its ligand (37). This may explain why epidermal, mammary, and prostate stem cells localize to the basal cell layer of their resident tissue, which directly contacts the basement membrane. The basement membrane and ECM can modulate and sequester the concentration of secreted factors within the stem cell niche and can act as scaffolds that effect signal transduction events (38). Proximity to such factors may be important in stem cell maintenance, because stem cell differentiation often occurs after departure from the basement membrane (39). Integrins also may play a more direct role in stem cell homeostasis, because they themselves are involved in signal transduction. Integrin binding with ligand leads to activation of signaling components including focal adhesive kinase and MAPK (38). Integrin ligation also has been reported to influence the synthesis of cytokines such as TGF-β, which is involved in regulating stem cell quiescence (40). Several studies have shown that the proximal region of the prostate exhibits a specialized architecture with a thick band of smooth muscle cells that produce high levels of TGF-β (41–43). CD49f therefore may not only mark prostate stem cells but also serve to anchor them within the niche and mediate contact with niche signals from the basement membrane and ECM.
Materials and Methods
Animals and Tissue Collection.
The β-actin GFP [C57BL/6-TgN(ACTbEGFP)1Osb], β-actin dsRED [Tg(ACTB-DsRed.MST)1Nagy/J], and SCID mouse strains were purchased from The Jackson Laboratory (Bar Harbor, ME). Eight- to 16-week-old C57BL/6 inbred mice originally purchased from The Jackson Laboratory were used for all experiments. Prostate cells were dissociated by mincing and collagenase digestion, and UGSM was harvested from embryonic day 16 fetuses as described in ref. 30.
Immunofluorescent and Histological Analysis.
Frozen prostate tissue sections and cytospins were prepared by air drying and fixation for 2 min in cold acetone as described in ref. 11. Tissue sections were incubated with primary antibodies diluted in 1× PBS at 4°C overnight. Sections subsequently were washed and incubated with secondary antibodies for 1 h at 25°C. Antibody sources and dilutions are listed in SI Materials. Sections were counterstained with DAPI (Vector Laboratories, Burlingame, CA) and analyzed by fluorescent microscopy.
Fluorescence-Activated Cell Sorting and Analysis.
Prostate cells were suspended in DMEM/10% FBS and stained with antibody for 20 min at 4°C. Antibody sources and dilutions are listed in SI Materials. FACS analysis was performed by using the BD FACS Canto and CellQuest software. Cell sorting was done by using the BD FACS Vantage (BD Biosciences).
In Vitro Prostate Colony- and Sphere-Forming Assays.
Colony assays were based on protocols in ref. 19. 3T3 cells (5 × 104) were plated in DMEM/10%FBS in each well of a six-well plate and irradiated with 500 rad the next morning. Prostate cell samples were counted by hemocytometer and plated in PrEGM (Cambrex, Walkersville, MD) on top of irradiated 3T3 cells. Colonies were counted on days 8–10. In some experiments, plates were fixed with acetone for 2 min, washed with 1× PBS, and stained with trypan blue for 1 h.
Prostate sphere growth and passaging conditions were based on protocols in refs. 23, 24, and 27). Each sample of prostate cells was counted by hemocytometer, mixed with 1 × 104 UGSM cells, and suspended in 1:1 Matrigel/PrEGM (BD Biosciences, Bedford, MA) in a total volume of 80 μl. Each sample was plated around the rim of a well of a 12-well plate and allowed to solidify for 15 min before 2 ml of PrEGM was added. Spheres were counted 7–10 days after plating. For passaging of spheres, media was aspirated and Matrigel was digested by incubation in 500 μl of dispase (Becton Dickinson, San Diego, CA) for 30 min at 37°C. Digested cultures were pelleted and incubated in 1 ml of PrEGM containing 10% collagenase for 30 min at 37°C. Samples again were pelleted and incubated in 0.05% Trypsin/EDTA for 10 min at room temperature, passed several times through a 27-gauge syringe, and passed over a 40-μm filter. Cells were counted by hemocytometer and replated at a density of 10,000 cells per well after each passage.
In Vivo Prostate Regeneration.
Prostate cells were counted by hemocytometer and mixed with 2 × 105 UGSM cells. Mixtures were pelleted and resuspended in 40 μl of Matrigel and kept on ice. Samples were injected s.c. on the backs of 8- to 16-week-old SCID mice by using an insulin syringe. Grafts were harvested five to eight weeks later. Serial tissue sections were stained with H&E to identify and count tubule structures.
Supplementary Material
Acknowledgments
We thank Connie Eaves, Robert Signer, Encarnacion Montecino-Rodriguez, and Caius Radu for advice on project design and FACS sorting and analysis. This work was supported by funds from the Prostate Cancer Foundation and the University of California, Los Angeles Special Program of Research Excellence in Prostate Cancer, (Jean deKernion, principal investigator). O.N.W. is an Investigator of the Howard Hughes Medical Institute. D.A.L. is supported by National Institutes of Health/Public Health Services Grant T32 CA09056 Tumor Cell Biology Training Grant.
Abbreviations
- CD
cluster designation
- CK
cytokeratin
- LSC
Lin−Sca-1+CD49f+
- SMA
smooth muscle actin
- UGSM
urogenital sinus mesenchyme.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609684104/DC1.
References
- 1.Spangrude GJ, Heimfeld S, Weissman IL. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 2.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 3.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 4.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 5.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat M-L, Wu L, Lindeman GJ, Visvader JE. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 6.Isaacs JT. In: Benign Prostatic Hyperplasia. Rodgers CH, Coffey DS, Cunha GR, editors. Vol 2. Washington, DC: U.S. Depart Health Hum Services; 1985. pp. 85–94. [Google Scholar]
- 7.Bhatt RI, Brown MD, Hart CA, Gilmore P, Ramani VA, George NJ, Clarke NW. Cytometry A. 2003;54:89–99. doi: 10.1002/cyto.a.10058. [DOI] [PubMed] [Google Scholar]
- 8.Tsujimura A, Koikawa Y, Salm S, Takao T, Coetzee S, Moscatelli D, Shapiro E, Lepor H, Sun TT, Wilson EL. J Cell Biol. 2002;157:1257–1265. doi: 10.1083/jcb.200202067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ, Collins AT. J Cell Sci. 2004;117:3539–3545. doi: 10.1242/jcs.01222. [DOI] [PubMed] [Google Scholar]
- 10.Burger PE, Xiong X, Coetzee S, Salm SN, Moscatelli D, Goto K, Wilson EL. Proc Natl Acad Sci USA. 2005;102:7180–7185. doi: 10.1073/pnas.0502761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xin L, Lawson DA, Witte ON. Proc Natl Acad Sci USA. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawson DA, Xin L, Lukacs R, Xu Q, Cheng D, Witte ON. Cold Spring Harb Symp Quant Biol. 2005;70:187–196. doi: 10.1101/sqb.2005.70.003. [DOI] [PubMed] [Google Scholar]
- 13.Liu AY, True LD. Am J Pathol. 2002;160:37–43. doi: 10.1016/S0002-9440(10)64346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. Breast Cancer Res. 2006;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Natali PG, Nicotra MR, Botti C, Mottolese M, Bigotti A, Segatto O. Br J Cancer. 1992;66:318–322. doi: 10.1038/bjc.1992.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter WG, Kaur P, Gil SG, Gahr PJ, Wayner EA. J Cell Biol. 1990;111:3141–3154. doi: 10.1083/jcb.111.6.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy RR, Hayakawa K. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 18.Sonnenberg A, Linders CJ, Daams JH, Kennel SJ. J Cell Sci. 1990;96:207–217. doi: 10.1242/jcs.96.2.207. [DOI] [PubMed] [Google Scholar]
- 19.Hudson DL, O'Hare M, Watt FM, Masters JR. Lab Invest. 2000;80:1243–1250. doi: 10.1038/labinvest.3780132. [DOI] [PubMed] [Google Scholar]
- 20.Uzgare AR, Xu Y, Isaacs JT. J Cell Biochem. 2004;91:196–205. doi: 10.1002/jcb.10764. [DOI] [PubMed] [Google Scholar]
- 21.English HF, Santen RJ, Isaacs JT. Prostate. 1987;11:229–242. doi: 10.1002/pros.2990110304. [DOI] [PubMed] [Google Scholar]
- 22.Goto K, Salm SN, Coetzee S, Xiong X, Burger PE, Shapiro E, Lepor H, Moscatelli D, Wilson EL. Stem Cells. 2006;24:1859–1868. doi: 10.1634/stemcells.2005-0585. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds BA, Weiss S. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 24.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webber MM, Bello D, Kleinman HK, Hoffman MP. Carcinogenesis. 1997;18:1225–1231. doi: 10.1093/carcin/18.6.1225. [DOI] [PubMed] [Google Scholar]
- 26.Lang SH, Sharrard RM, Stark M, Villette JM, Maitland NJ. Br J Cancer. 2001;85:590–599. doi: 10.1054/bjoc.2001.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bello-DeOcampo D, Kleinman HK, Deocampo ND, Webber MM. Prostate. 2001;46:142–153. doi: 10.1002/1097-0045(20010201)46:2<142::aid-pros1018>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 28.Singec I, Knoth R, Meyer RP, Maciaczyk J, Volk B, Nikkhah G, Frotscher M, Snyder EY. Nat Methods. 2006;3:801–806. doi: 10.1038/nmeth926. [DOI] [PubMed] [Google Scholar]
- 29.Cunha GR, Lung B. J Exp Zool. 1978;205:181–193. doi: 10.1002/jez.1402050203. [DOI] [PubMed] [Google Scholar]
- 30.Xin L, Ide H, Kim Y, Dubey P, Witte ON. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins AT, Habib FK, Maitland NJ, Neal DE. J Cell Sci. 2001;114:3865–3872. doi: 10.1242/jcs.114.21.3865. [DOI] [PubMed] [Google Scholar]
- 32.Azuma M, Hirao A, Takubo K, Hamaguchi I, Kitamura T, Suda T. Biochem Biophys Res Commun. 2005;338:1164–1170. doi: 10.1016/j.bbrc.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 33.Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- 34.Tani H, Morris RJ, Kaur P. Proc Natl Acad Sci USA. 2000;97:10960–10965. doi: 10.1073/pnas.97.20.10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones PH, Watt FM. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]
- 36.Fortunel NO, Otu HH, Ng HH, Chen J, Mu X, Chevassut T, Li X, Joseph M, Bailey C, Hatzfeld JA, et al. Science. 2003;302:393. doi: 10.1126/science.1086384. author reply 393. [DOI] [PubMed] [Google Scholar]
- 37.Colognato H, Yurchenco PD. Dev Dyn. 2000;218:213–234. doi: 10.1002/(SICI)1097-0177(200006)218:2<213::AID-DVDY1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 38.Juliano RL. Annu Rev Pharmacol Toxicol. 2002;42:283–323. doi: 10.1146/annurev.pharmtox.42.090401.151133. [DOI] [PubMed] [Google Scholar]
- 39.Jensen UB, Lowell S, Watt FM. Development (Cambridge, UK) 1999;126:2409–2418. doi: 10.1242/dev.126.11.2409. [DOI] [PubMed] [Google Scholar]
- 40.Wang D, Sun L, Zborowska E, Willson JK, Gong J, Verraraghavan J, Brattain MG. J Biol Chem. 1999;274:12840–12847. doi: 10.1074/jbc.274.18.12840. [DOI] [PubMed] [Google Scholar]
- 41.Nemeth JA, Lee C. Prostate. 1996;28:124–128. doi: 10.1002/(SICI)1097-0045(199602)28:2<124::AID-PROS8>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 42.Nemeth JA, Sensibar JA, White RR, Zelner DJ, Kim IY, Lee C. Prostate. 1997;33:64–71. doi: 10.1002/(sici)1097-0045(19970915)33:1<64::aid-pros11>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 43.Salm SN, Burger PE, Coetzee S, Goto K, Moscatelli D, Wilson EL. J Cell Biol. 2005;170:81–90. doi: 10.1083/jcb.200412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.