Abstract
We examined the role of hepatic heparan sulfate in triglyceride-rich lipoprotein metabolism by inactivating the biosynthetic gene GlcNAc N-deacetylase/N-sulfotransferase 1 (Ndst1) in hepatocytes using the Cre-loxP system, which resulted in an approximately 50% reduction in sulfation of liver heparan sulfate. Mice were viable and healthy, but they accumulated triglyceride-rich lipoprotein particles containing apoB-100, apoB-48, apoE, and apoCI-IV. Compounding the mutation with LDL receptor deficiency caused enhanced accumulation of both cholesterol- and triglyceride-rich particles compared with mice lacking only LDL receptors, suggesting that heparan sulfate participates in the clearance of cholesterol-rich lipoproteins as well. Mutant mice synthesized VLDL normally but showed reduced plasma clearance of human VLDL and a corresponding reduction in hepatic VLDL uptake. Retinyl ester excursion studies revealed that clearance of intestinally derived lipoproteins also depended on hepatocyte heparan sulfate. These findings show that under normal physiological conditions, hepatic heparan sulfate proteoglycans play a crucial role in the clearance of both intestinally derived and hepatic lipoprotein particles.
Introduction
Heparan sulfate proteoglycans (HSPGs) act as receptors for growth factors, chemokines, enzymes, and cell adhesion proteins, thereby modulating their concentration, distribution and biological activity (1–3). Additionally, membrane-bound proteoglycans can act as endocytic receptors facilitating the uptake of various ligands and metabolites (4, 5). The heparan sulfate (HS) side chains assemble by the alternating addition of N-acetyl- d-glucosamine (GlcNAc) and d-glucuronic acid units to a tetrasaccharide primer linked to specific serine residues in the core proteins of the proteoglycans (for a review, see ref. 3). During polymerization, a series of modification reactions occur by processing enzymes located in the Golgi apparatus. A family of 4 GlcNAc N-deacetylase/N-sulfotransferases (Ndsts) removes acetyl groups from subsets of GlcNAc residues and adds sulfate to the free amino groups. An epimerase converts adjacent d-glucuronic acid units to l-iduronic acid, and then a group of O-sulfotransferases add sulfate to C6 and C3 of glucosamine units and C2 of uronic acids. These modifications occur in contiguous blocks of sugars along the chain in an incomplete manner, resulting in domains of variable size and sulfation. Sets of modified sugars act as binding sites for different proteins, often with great specificity and affinity (1).
Exogenous lipids enter the circulation as large, buoyant chylomicrons, which arise in intestinal enterocytes and contain apoB-48. Endogenous lipids made in the liver appear in the circulation as VLDLs and contain apoB-100 (and apoB-48 in mice) synthesized by hepatocytes. As these triglyceride-rich lipoproteins (TRLs) circulate, lipoprotein lipase (Lpl) hydrolyzes the triglycerides in the cores of the particles, resulting in the formation of chylomicron remnants (CRs) and intermediate-density lipoproteins (IDLs) from VLDLs. Lpl also can remain associated with lipoprotein particles (6), although most of the enzyme associates with HSPGs on the surface of endothelial cells (7). TRL remnants also obtain apoE by transfer reactions from other lipoproteins in the circulation or by enrichment in the liver. apoB-48, apoB-100, apoE, and Lp1, have in common the capacity to bind HS.
The mechanism of hepatic clearance of TRLs remains controversial. The particles first may be sequestered in the liver perisinusoidal space (space of Disse), where they undergo further processing by hepatic lipase (HL) and Lpl, both of which are detectable in the space of Disse and may remain associated with TRLs (8–10). A group of hepatic lipoprotein receptors then endocytoses the lipoproteins, leading to their lysosomal catabolism. Multiple receptors mediating clearance of TRL remnants appear to exist. Clearance through the LDL receptor (LDLR) was hypothesized to be the primary mechanism based on its ability to recognize apoB-100 and apoE. However, clearance of TRLs occurs normally in humans, WHHL rabbits, and mice deficient in LDLR (11–13). Thus, early on it was recognized that an LDLR-independent remnant receptor must exist. After the identification of several false candidates, the LDLR-related protein (LRP) scavenger receptors were shown to bind and internalize remnant particles (14). However, inactivation of LRP1 or LRP5 alone does not result in remnant particle accumulation in animals fed normal chow (i.e., under normal physiological conditions), bringing the role of LRPs into question (15, 16). Combining LRP1 with LDLR mutations results in only a modest accumulation of TRLs (15).
Cell-surface HSPGs have been hypothesized to act as a third class of remnant receptors acting either independently or in concert with LRP (6, 17–26). In cell culture models, HSPGs can interact directly with lipoproteins bearing apoE, Lpl, and HL and facilitate their internalization by endocytosis (6, 17, 23). All 3 proteins bind to heparin in vitro and HS on cultured cells dependent on sulfation of the chains (27–32). As hepatocyte HS is notably more enriched in sulfate than HS from endothelium and other tissues (27, 33, 34), binding of these lipoproteins to liver HS may be preferred to binding to HS in other tissues. Infusion of mice with heparinase, heparin, suramin, or lactoferrin to neutralize HS reduced plasma clearance rates and reduced hepatic uptake of labeled VLDL (35–37), suggesting that the interaction of particles with HS could be important in vivo. Several rare apoE alleles exhibit reduced binding to HS and cause severe hyperlipoproteinemia (38, 39), and mice bearing Lpl defective in HS binding fail to traffic lipids to tissues properly (40). Together, these findings suggest a role for HS in clearance of TRLs, but direct demonstration of HS-mediated clearance of remnant particles in vivo has not been demonstrated.
In the current study, we have genetically tested whether HS plays a role in lipoprotein metabolism in vivo by altering sulfation of the chains specifically in hepatocytes and endothelial cells using the Cre-loxP system. Decreasing sulfation of HS in hepatocytes in this way resulted in the accumulation of VLDL-like particles under normal physiological conditions, CRs after oral gavage of triglycerides, and cholesterol-rich particles in animals lacking LDLRs. Thus, proteoglycans containing HS constitute an essential component for clearance of TRLs and may represent the long-sought-after receptors for remnant particles.
Results
Hepatocyte-specific Ndst1 gene disruption alters HS synthesis in liver.
Liver contains transcripts for 2 isozymes of GlcNAc N-deacetylase/N-sulfotransferase, Ndst1 and Ndst2. Systemic inactivation of Ndst1 in mice causes perinatal lethality due to brain and lung defects (41–43), and Ndst1–/– embryos exhibit altered liver HS structure (41, 44). In contrast, Ndst2–/– mice are viable and do not exhibit any changes in liver HS structure in spite of a high level of enzyme expression in liver (44–46). To study the role of Ndst1 in liver physiology, we crossbred mice bearing a conditional loxP-flanked (“floxed”) allele of Ndst1 (Ndst1f/f) (47) to transgenic mice expressing Cre recombinase under control of the rat albumin promoter (AlbCre) to drive inactivation of the gene (48). Expression of the AlbCre transgene occurs after postnatal day 10 only in hepatocytes. From these breedings, we established a colony of hepatocyte-specific Ndst1-knockout (Ndst1f/fAlbCre+) mice with littermate controls of the genotype Ndst1f/fAlbCre–. All genotypes were congenic and maintained on a C57BL/6J background. They were viable and fertile, yielding litters of normal size and the expected Mendelian ratio of genotypes. Adult livers were morphologically normal by histology, and serum levels of alkaline phosphatase, albumin, and aspartate and alanine aminotransferases indicated no overt liver pathophysiology (data not shown).
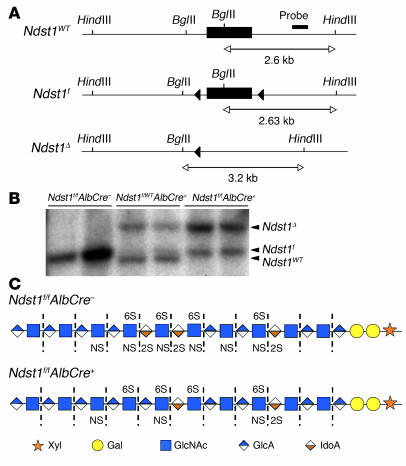
Hepatocytes were isolated from adult mice to measure the extent of Ndst1 inactivation by Southern blotting (Figure 1A). Cre-mediated recombination deleted the loxP-flanked exon, removing a BglII restriction site and yielding a BglII/HindIII fragment of 3.2 kb, whereas the wild-type allele generated a 2.6-kb fragment. DNA from hepatocytes of 8-week-old heterozygous mice (Ndst1f/WTAlbCre+) showed both the wild-type allele (slightly smaller than the 2.63-kb BglII/HindIII fragment of the Ndst1f allele) and incomplete recombination of the Ndst1f allele (Figure 1B). By scanning the bands, we concluded that 65%–75% of the Ndst1f alleles had undergone recombination in adult mice. Homozygous (Ndst1f/fAlbCre+) mice showed a similar extent of inactivation, indicating that negative selection against Ndst1-deleted hepatocytes did not occur. The extent of recombination in animals increased up to 6 weeks of age (data not shown), in agreement with the behavior of other loxP-targeted genes inactivated by AlbCre expression (48).
Figure 1. Ndst1 conditional knockout.
(A) Gene schematic and Southern blotting strategy for mapping Ndst1 alleles. Filled triangles represent loxP recombination sites; bar indicates target sequence of Ndst1 probe; restriction sites for BglII and HindIII and expected fragment sizes are also indicated. (B) Southern blot to determine Cre-mediated recombination of Ndst1 floxed alleles. Hepatocyte genomic DNA was digested with BglII/HindIII and probed for Ndst1 alleles. The deleted allele gave a BglII/HindIII fragment at 3.2 kb, and the Ndst1f and wild-type alleles gave BglII/HindIII fragments at 2.6 kb. The Ndst1f BglII/HindIII fragment was slightly larger than 2.6 kb due to the inserted 34-bp loxP site. DNA in each lane was isolated from individual mice. Quantification of bands indicated 65%–75% recombination in AlbCre+ hepatocytes. (C) The HS chains are depicted using the indicated symbol nomenclature for the individual sugar residues. Heparinases degrade the chains to disaccharides (dashed lines), which can then be separated according the number and pattern of sulfate groups (Table 1). HS chains from the mutant contain less sulfate and iduronic acid because Ndst1 creates the preferred substrates for epimerization and O-sulfation. The residual sulfation presumably arises from incomplete inactivation of Ndst1 and the expression of Ndst2.
Although Ndst1 inactivation was incomplete, the mutation induced significant changes in liver HS composition. HS was isolated from whole livers of adult mice (n = 5 per genotype) and depolymerized into its constituent disaccharides by bacterial heparinases (Figure 1C). The disaccharides were separated by HPLC and quantitated by post-column derivatization and fluorometry (Table 1). Levels of disaccharides containing N-sulfated glucosamine residues (i.e., unsaturated uronic acid–N-sulfo- d-glucosamine [ΔUA-GlcNS], ΔUA-GlcNS6S, ΔUA2S-GlcNS, and ΔUA2S-GlcNS6S) were reduced from 39% in HS from wild-type livers to 18% in the mutant. The 2-O-sulfation of uronic acids (ΔUA2S-GlcNS and ΔUA2S-GlcNS6S) showed a larger decrease (15% in controls to 3% in the mutant), consistent with previous analyses of HS from mice containing systemic inactivation of Ndst1 (43, 44). In contrast, glucosamine 6-O-sulfation (ΔUA-GlcNAc6S, ΔUA-GlcNS6S, and ΔUA2S-GlcNS6S) remained unchanged (21% of total disaccharides in control mice and 23% in mutant mice). Since the presence of endothelial, stellate, and Kupffer cells in whole liver might have obscured some of the effects of Ndst1 inactivation on hepatocyte HS, we examined the disaccharides derived from HS from highly purified hepatocytes (Table 1). Although the composition differed somewhat from that of whole liver HS, inactivation of Ndst1 resulted in similar effects on N-, 2-O-, and 6-O-sulfation. Levels of N-sulfated disaccharides declined from 38% to 18%; 2-O-sulfated disaccharides were reduced from 17% to 4%; and 6-O-sulfated disaccharides remained relatively unaffected (31% versus 26%).
Table 1 .
Disaccharide analysis of liver HS
Ndst1f/fAlbCre+ mice accumulate TRLs.
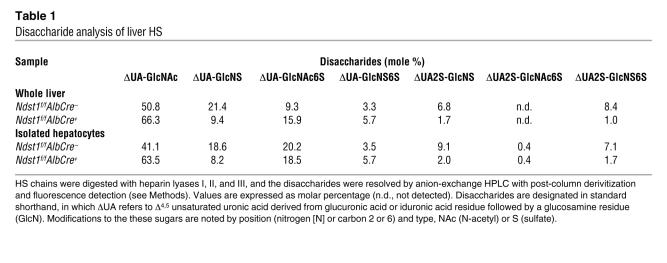
Analysis of plasma samples from overnight-fasted 8-week-old mice showed that total plasma triglyceride levels were approximately 2.5-fold higher in Ndst1-deficient mice (36.8 ± 10.4 mg/dl in control mice versus 91.7 ± 35.0 mg/dl; n = 20; P < 0.0001) (Figure 2A). Greater variation in plasma triglyceride levels was observed in mutant mice compared with the controls, possibly due to the variable inactivation of Ndst1 in individual animals. Total plasma cholesterol was not affected (80.1 ± 10.1 mg/dl versus 81.4 ± 9.1; n = 20; P = 0.68) (Figure 2B), which contrasts the phenotype of LDLR and LRP mutant mice (15, 49). Triglyceride accumulation persisted in adult mice regardless of age (2–12 months) and showed the same level of variation.
Figure 2. Hypertriglyceridemia in Ndst1-deficient mice.
Total triglycerides (A) and total cholesterol (B) were measured in plasma samples (n = 20 per genotype, mixed males and females, horizontal bars indicate mean values). Triglyceride (C) and cholesterol (D) content of lipoproteins fractionated by preparative density ultracentrifugation was determined. Each value is from 1 set of pooled plasma derived from 10 mice. Lipoproteins were also analyzed by FPLC gel filtration, and the amount of triglyceride (E) and cholesterol (F) was determined in control mice (filled circles) and Ndst1f/fAlbCre+ mice (open circles). Plasma was pooled from 9 mice of each genotype. The elution positions of human VLDL, IDL/LDL, and HDL are indicated. (G) Samples of purified lipoproteins (d < 1.019 g/ml) were analyzed by gradient SDS-PAGE, and the individual apolipoproteins were visualized by Coomassie blue or silver staining. The location of apoB-48, apoB-100, apoE, and apoAI was determined by Western blotting (data not shown). The location of apoCs and serum albumin was deduced by Mr values.
Density ultracentrifugation of pooled plasma samples showed that Ndst1-deficient mice accumulated triglyceride-rich particles in the CR, VLDL, and IDL density range (Figure 2C). No significant change was noted in either LDL or HDL. Cholesterol distribution among the different lipoprotein density classes remained essentially unchanged (Figure 2D), supporting the results of plasma analyses (Figure 2B). Lipoprotein profiles determined by gel filtration fast-phase liquid chromatography (FPLC) also documented the accumulation of large TRLs consistent in size with CR, VLDL, and IDL (Figure 2E), with no change in cholesterol distribution (Figure 2F).
SDS-PAGE analysis of the VLDL and IDL fractions obtained by ultracentrifugation (d < 1.019 g/ml) showed that the accumulated particles contained apoB-48, apoB-100, apoE, and various apoCs typical of TRLs (Figure 2G). Samples were delipidated and analyzed after trypsin digestion by mass spectrometry, which confirmed these findings and demonstrated the presence of apoAII, apoCI, apoCII, apoCIII, and apoCIV. apoAIV, apoAV, Lpl, and HL were not detected by SDS-PAGE analysis or mass spectrometry, suggesting that the particles that accumulated in the mutant were not enriched for these apolipoproteins or lipases or that they were lost during ultracentrifugation.
HSPGs mediate clearance of cholesterol-rich lipoproteins.
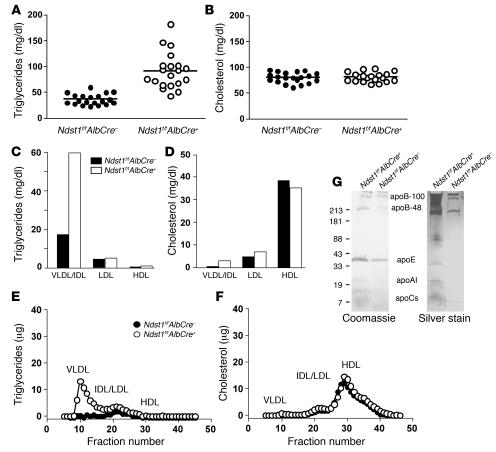
To examine the relative contribution of HS and LDLRs to lipoprotein catabolism, Ndst1f/fAlbCre+ mice were crossbred with Ldlr–/– mice. The compound mutants accumulated a large excess of plasma triglycerides compared with Ldlr–/– mice (323.0 ± 69.2 mg/dl versus 92.5 ± 25.1 mg/dl; n = 11; P < 0.0001) (Figure 3A). Higher plasma cholesterol levels were also detected in compound mutants compared with Ldlr–/– mice (379.8 ± 75.7 mg/dl versus 249.3 ± 22.0 mg/dl; n = 11; P < 0.0001) (Figure 3B). Ultracentrifugal flotation studies showed that the compound mutants accumulated particles of d < 1.019 g/ml rich in triglycerides and cholesterol (Figure 3, C and D). FPLC analysis documented marked increases in levels of triglyceride-rich VLDL and IDL (Figure 3E). As expected, Ldlr–/– mice displayed elevated LDL cholesterol levels, which was further accentuated in the double mutants (Figure 3F). In contrast, the content of HDL cholesterol was similar in the 4 genotypes. These findings suggested that hepatic HS acts in the clearance of cholesterol-rich lipoprotein particles as well as those carrying triglycerides, particularly when LDLRs are absent or downregulated. Alteration of liver HS structure did not affect expression levels of LDLR or LRP1 in the liver as measured by quantitative PCR (data not shown), and LDL metabolism appeared to be normal, since particles did not accumulate in Ndst1f/fAlbCre+ mice.
Figure 3. Cholesterol and triglyceride particles accumulated in Ldlr–/–Ndst1f/fAlbCre+ mice.
Total plasma triglyceride (A) and cholesterol (B) levels were determined for Ldlr–/–Ndst1f/fAlbCre– mice (filled circles) and Ldlr–/–Ndst1f/fAlbCre+ mice (open circles). Points represent individual animals; n = 11 per genotype; horizontal bars indicate mean values. All mice (8–10 weeks old) were fed normal chow and fasted overnight prior to analysis. Double mutants showed significant accumulation of both triglycerides and cholesterol. Triglyceride (C) and cholesterol (D) content of lipoproteins fractionated according to density by preparative ultracentrifugation. Each value is from 1 set of pooled plasma derived from 10 mice. Lipoproteins were analyzed by FPLC gel filtration, and the amount of triglyceride (E) and cholesterol (F) was determined in Ldlr–/–Ndst1f/fAlbCre– mice (filled circles) and Ldlr–/–Ndst1f/fAlbCre+ mice (open circles). Plasma was pooled from 10 mice of each genotype. The elution positions of human VLDL, IDL/LDL, and HDL are indicated.
Since the extent of Ndst1 inactivation in hepatocytes was only 65%–75% (Figure 1B), we wondered whether a more complete deletion of the gene would accentuate the hyperlipidemia in these animals. Therefore, we injected approximately 4 × 1011 particles of adenovirus Cre (AdCre) (49) via the tail vein into adult Ndst1f/fAlbCre–, Ndst1f/fAlbCre+, Ldlr–/–Ndst1f/fAlbCre–, and Ldlr–/–Ndst1f/fAlbCre+ mice and waited 3 weeks to allow turnover of preexisting HS chains. Southern blot analysis of hepatocyte DNA showed that AdCre treatment increased the extent of recombination to 80%–90% (data not shown). However, inactivation of Nsdt1 by AdCre increased fasting serum lipid levels to the same extent as inactivation of Ndst1 by AlbCre, and no additive effect was observed (e.g., triglyceride levels increased from 55.5 ± 10.2 mg/dl in Ldlr–/–Ndst1f/fAlbCre–AdCre– to 157.3 ± 38.9 mg/dl in Ldlr–/–Ndst1f/fAlbCre–AdCre+ mice compared with 147.2 ± 37.3 mg/dl in Ldlr–/–Ndst1f/fAlbCre+AdCre– and 135.7 ± 37.1 mg/dl in Ldlr–/–Ndst1f/fAlbCre+AdCre+ mice; n = 6). Thus, more complete inactivation of Ndst1 did not potentiate the phenotype.
HS-deficient mice exhibit slower plasma clearance of dietary triglycerides and VLDL.
The accumulation of TRLs in Ndst1-deficient mice could reflect increased secretion of lipoproteins or decreased clearance or reuptake of particles after secretion (51). As determined after injection of animals with Triton WR-1339 to inhibit lipolysis and lipoprotein clearance (52), triglyceride secretion rates did not differ significantly between Ndst1f/fAlbCre+ and control mice (0.055 ± 0.008 mg/min [n = 6] versus 0.047 ± 0.005 mg/min [n = 4], respectively; P = 0.4973). This finding indicates that the accumulation of triglycerides most likely results from decreased clearance or reuptake.
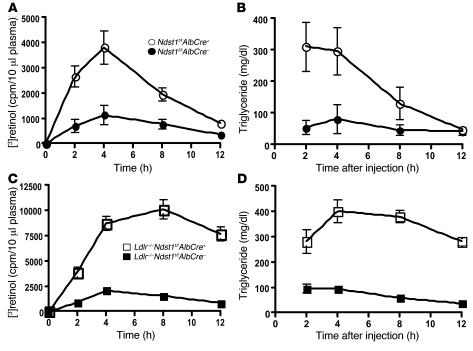
Clearance rates of intestinally derived TRLs were measured in mice lacking hepatocyte HS by a vitamin A fat tolerance test. Overnight-fasted mice were given a bolus mixture of [3H]retinol and corn oil by gavage, and blood was sampled at various time points from the retro-orbital sinus. Retinyl ester excursion curves revealed that Ndst1f/fAlbCre+ mice cleared intestinally derived lipoproteins at a slower rate than wild-type mice, but most intestinally derived particles were eventually cleared by 12 hours (Figure 4A). Ldlr–/–Ndst1f/fAlbCre+ mice metabolized intestinally derived TRLs at an even slower rate, with less than 25% of the counts cleared after 12 hours (Figure 4C), indicating that these 2 receptors work in concert to clear dietary triglyceride. Plasma triglyceride clearance matched the retinal ester excursion curves in all 4 genotypes (Figure 4, B and D). Analysis of plasma samples by density ultracentrifugation showed that the majority of the counts and triglycerides were in lipoprotein particles with d < 1.006 g/ml (data not shown).
Figure 4. Slowed clearance of postprandial lipoproteins.
Retinyl ester excursions and plasma triglyceride levels were measured at the times indicated in 8-week-old Ndst1f/fAlbCre+ mice (n = 9) compared with wild-type siblings (n = 5) (A and B) and Ldlr–/–Ndst1f/fAlbCre+ mice (n = 7) compared with Ldlr–/–Ndst1f/fAlbCre– siblings (n = 4) (C and D). Overnight-fasted animals of the indicated genotypes were given 200 μl of corn oil containing 5.4 μCi [3H]retinol by gavage. Blood samples were taken at the indicated times, and radioactivity remaining in 10 μl of serum was determined by scintillation counting in triplicate. The values are expressed as mean ± SD.
Clearance of VLDL was also affected based on experiments in which a bolus of human VLDL was injected intravenously and the circulating level of human VLDL was determined by ELISA using mAb MB47, which is specific for human apoB-100 (53) (Figure 5A). The rate of turnover was reduced in mutant mice, yielding apparent half-lives (calculated by extrapolation from the curves) of 23.2 ± 1.4 minutes in control mice and 46.6 ± 5.3 minutes in mutant mice (n = 6 per genotype; P = 0.0016). Thus, hepatocyte HS plays a central role in the clearance of both intestinally and hepatically derived lipoprotein particles.
Figure 5. Decreased plasma clearance and liver uptake of VLDL in Ndst1-deficient mice.
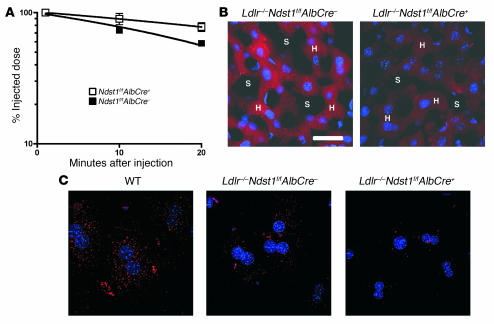
(A) Plasma clearance of human VLDL apoB-100 was measured by ELISA using human apoB-100–specific monoclonal antibody MB47 (see Methods). Control mice (filled squares; t1/2 = 23.2 ± 1.4 minutes; n = 6) and Ndst1f/fAlbCre+ mice (open squares; t1/2 = 46.6 ± 5.3 minutes; n = 6). The difference in t1/2 between the genotypes was significant (P = 0.0016), and the difference between the 10- and 20-minute time points for the mutant was also significant (P = 0.0133). Results were verified in a separate experiment. (B) Deconvolution microscopy showing uptake of DiI-VLDL (red) in the liver of Ldlr–/–Ndst1f/fAlbCre– and Ldlr–/–Ndst1f/fAlbCre+ mice. DiI-VLDL (100 μg) was injected by tail vein, and livers were fixed 20 minutes later. Sections of 20 μm were DAPI-stained for nuclei (blue). H, hepatocyte cords; S, sinusoids. Scale bar: 26 μm; magnification, ×40. (C) Hepatocytes were isolated from mice of the indicated genotypes and incubated with DiI-VLDL (5 μg/ml). Images were obtained by deconvolution microscopy.
We also examined hepatic uptake of lipoproteins in vivo by injecting DiI-labeled human VLDL into Ndst1 mutant and wild-type mice. After 20 minutes, livers were fixed, sectioned, and examined by deconvolution fluorescence microscopy. Initial studies did not reveal any hepatic staining, presumably owing to rapid disposal of labeled particles by clearance via LDLR-mediated endocytosis. When the experiment was repeated in Ldlr–/– mice, pericellular staining of hepatocytes was observed with accumulation of DiI-VLDL in intracellular vesicles, indicating particles were bound and taken up by the hepatocytes (Figure 5B). Considerably less DiI-VLDL was retained by the hepatocytes in Ldlr–/–Ndst1f/fAlbCre+ mice. Defective binding and uptake of DiI-VLDL was also observed in isolated hepatocytes from Ldlr–/–Ndst1f/fAlbCre+ mice compared with Ldlr–/–Ndst1f/fAlbCre– or wild-type hepatocytes (Figure 5C), indicating a cell-autonomous effect on clearance.
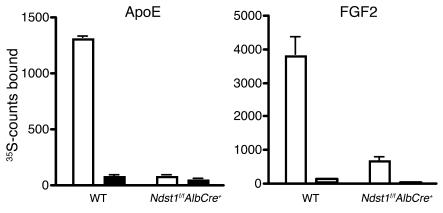
Based on these studies, we predicted that the reduction in sulfation of HS in Ndst1-deficient mice should reduce binding to apoE/apoB-bearing particles to isolated HS. To test this hypothesis, radiolabeled HS was isolated from cultured hepatocytes derived from Ndst1f/fAlbCre+ and wild-type mice and incubated with recombinant apoE. Bound chains were collected by rapid filtration on a membrane that binds HS complexed to protein but not free chains. As shown in Figure 6, apoE bound HS derived from wild-type cells, and the interaction was inhibited by inclusion of heparin in the incubation medium. In contrast, binding of chains derived from the mutant was dramatically reduced, and added heparin had little additional effect. The extent of binding to FGF2, a growth factor that interacts with high affinity with HS, was also reduced (54).
Figure 6. Altered binding of mutant HS to apoE and FGF2.
Radiolabeled HS was isolated from cultured primary hepatocytes from Ndst1f/fAlbCre+ mice or wild-type controls grown in the presence of 35SO4 (see Methods). Samples (104 cpm) were incubated in physiologic saline with 10 μg recombinant apoE (lipid-free) or FGF2 in the absence (white bars) and presence (black bars) of 100 μg/ml of heparin, and bound material was collected by membrane filtration. Each experiment was done in triplicate, and the average values are shown.
Elevated levels of HL circulate in Ndst1f/fAlbCre+ mice.
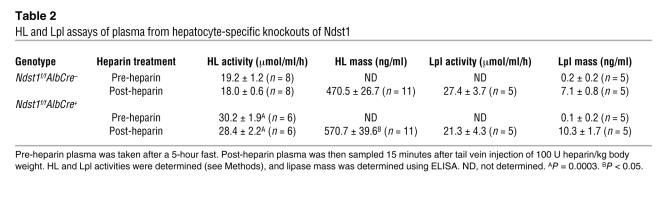
Both HL and Lpl have been implicated in assisting hepatic lipoprotein endocytosis, in some cases independent of enzymatic activity (55–57). Because these enzymes also bind HS (29–32), we tested plasma samples for changes in lipase activities before and after heparin injection (Table 2). Both mass and enzymatic activity of HL were elevated in Ndst1f/fAlbCre+ plasma after heparin treatment. Most of the HL was freely circulating in both mutant and control mice, since the levels did not increase measurably by heparin displacement. In contrast, very little Lpl was present in the circulation in both mutant and wild-type mice. Heparin injection dramatically increased the level of enzyme to comparable levels in both mutant and wild-type mice.
Table 2 .
HL and Lpl assays of plasma from hepatocyte-specific knockouts of Ndst1
Inactivation of endothelial cell Ndst1 has no effect on plasma lipoprotein metabolism.
Since endothelial cells that surround the liver sinusoids also express proteoglycans, we inactivated Ndst1 in these cells by crossing Ndst1f/f animals with TekCre mice to drive expression of the Cre recombinase in endothelial cells as well as in myeloid and lymphoid cells (47). Crossbreeding Ndst1f/f mice with TekCre mice resulted in greater than 95% inactivation of the gene in endothelial cells derived from the liver or lung microvasculature (47). Altering Ndst1 in these cells also led to changes in HS composition similar to those observed in hepatocytes, except that 2-O-sulfation was not reduced as dramatically (47). These changes in HS led to reduced leukocyte extravasation due to altered binding of L-selectin and chemokines to the endothelium (47). However, analysis of plasma samples showed that altering HS synthesis in endothelial cells had no effect on plasma triglycerides (95 ± 10 mg/dl in Ndst1f/fTekCre+ mice versus 83 ± 5 mg/dl in Ndst1f/fTekCre– mice; n = 17–21; P > 0.05) or cholesterol (76 ± 5 versus 79 ± 4 mg/dl, respectively).
Discussion
In this study, we have established that hepatocyte HS plays a crucial role in lipoprotein metabolism independent of LDLR family members. The major findings include the following observations: (a) Altering the fine structure of HS selectively in hepatocytes causes hyperlipoproteinemia but not overt liver pathophysiology; (b) Ndst1 deficiency causes accumulation of large TRLs resembling remnant particles derived from VLDL and chylomicrons; (c) Compounding the mutation with LDLR deficiency results in the additional accumulation of cholesterol-rich particles resembling IDL/LDL; (d) The hyperlipoproteinemia is linked to defective clearance of both postprandial and hepatic derived particles by the liver; (e) HL accumulates in the plasma; and (f) A similar alteration of HS structure in endothelial cells has no effect on lipoprotein metabolism. These findings suggest that hepatocyte proteoglycans bearing HS represent the long-sought-after receptors for remnant lipoprotein clearance in the liver.
HSPGs mediate the clearance of multiple lipoprotein subclasses.
Ndst1-deficient mice differ in phenotype from strains altered in classical lipoprotein receptors (LDLR and LRP), revealing the unique function of proteoglycans in liver-mediated clearance. LDLR deficiency leads to a large increase in levels of apoB-100–bearing cholesterol-rich LDL and an increase in the level of triglyceride-rich particles (49), whereas inactivation of LRP1 in the liver (15) or systemic inactivation of LRP5 has no effect on lipoprotein clearance under normal physiological conditions (58). Compounding deficiencies in LRP1 and LDLR results in an increase in the level of apoB48/apoE particles and a 2-fold increase in plasma cholesterol and triglyceride levels (15), suggesting that LRP1 can aid in the metabolism of lipoproteins, but its activity is masked by LDLR under normal conditions. In contrast, inactivation of Ndst1 in the liver resulted in accumulation of a distinct set of lipoproteins, containing apoB-100, apoB-48, and apoE and rich in triglycerides but low in cholesterol. Compound mutants lacking both Ndst1 and Ldlr accumulated high levels of triglyceride-rich particles (3.5-fold increase compared with LDLR deficiency alone) as well as cholesterol-rich lipoproteins IDL and LDL (1.6-fold compared with LDLR deficiency alone; Figure 3, A and B). Thus, hepatocyte HS plays a central role in the metabolism of all subclasses of lipoproteins, with the exception of HDL, as suggested previously (25). Additional studies are underway to address the effect of compounding Ndst1 and Lrp mutations and whether changes in HS in extrahepatic tissues might affect lipoprotein metabolism as well.
Metabolism of dietary triglycerides also appears to require liver HS. Earlier studies established that 25%–50% of CR particles are cleared by LDLR, depending on the methods used (13). Evidence exists that LRP can clear CR as well, but the failure of α-macroglobulin, a ligand for LRP, to compete with lipoprotein binding to cells casts doubt on this idea. Multiple studies pointed to a third receptor sensitive to heparinase, heparin, suramin, and lactoferrin (35–37), suggesting that the interaction of particles with HS could be important in vivo. Our results provide direct in vivo evidence supporting a role for HS in clearance of postprandial lipoproteins and indicate that this mechanism plays a more significant but overlapping role with LDLR (compare Ldlr–/–Ndst1f/fAlbCre– with Ndst1f/fAlbCre+ mice; Figure 4). HS and LDLR represent parallel systems for CR clearance, since the long delay in removal of dietary triglycerides in compound mutants mimics the delay seen in apoE-deficient mice (13).
HSPGs may act through multiple mechanisms.
Several mechanisms have been suggested to explain how HS-mediated interactions with apolipoproteins and enzymes affect lipoprotein processing in the liver. Mahley and coworkers have advanced the idea that remnant particles entering the space of Disse bind to HSPGs via apoE, resulting in their sequestration prior to further processing and receptor-mediated endocytosis (8) (Figure 7). Lpl and HL can also act in this way, since both lipases bind to HS and to the particles (4, 8) and have been localized to the space of Disse by immunoelectron microscopy (9, 10). Thus, one way to explain the accumulation of lipoproteins in the Ndst1 mutant is that reduced sulfation of the chains decreases sequestration of remnant particles, which can then pass back out of the space of Disse into the circulation. Reduced sulfation of heparan chains could also decrease the pool of apoE/Lpl/HL associated with the hepatocyte plasma membrane (9, 10, 59), which may act as a reservoir for enriching TRLs with these ligands or as molecular bridges between lipoproteins their receptors.
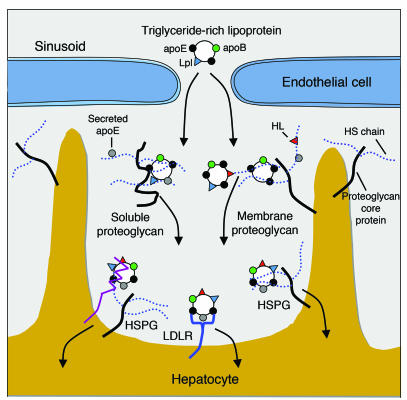
Figure 7. Model of possible roles for hepatic HS in TRL clearance.
Hepatocytes and endothelial cells produce membrane-bound HSPGs and secrete proteoglycans into the space of Disse. After lipolytic processing of lipoproteins in the circulation by Lpl (blue triangles), apoE-enriched (black circles) remnant lipoproteins enter the space of Disse through fenestrations in the endothelium. Remnant lipoproteins are thought to be sequestered near the hepatocyte cell surface via apoE-HS binding or lipase-HS bridging on secreted HSPGs. Lipoproteins are further processed in the space of Disse by transfer of soluble apoE (gray circles) and by HL (red triangles) bound via HS. apoE, HL, and Lpl can potentially serve as ligands of TRLs. Endocytosis of lipoprotein particles occurs via LDLR (blue) or LRP (purple) in association with HSPGs or independently by proteoglycans. Adapted with permission from Journal of Lipid Research (8) and Arteriosclerosis, Thrombosis, and Vascular Biology (86).
At first glance, the increase in the level of plasma HL in Ndst1f/fAlbCre+ mice is consistent with the idea that the decrease in clearance could reflect mislocalization of the enzyme. However, we believe this explanation is unlikely, since genetic inactivation of HL in mice results in accumulation of HDL, not TRL remnants (60). The increase in the level of plasma HL supports the idea that a portion of the enzyme is normally sequestered in the liver by way of HS and may be cleared in this way by endocytosis. The lack of plasma accumulation of Lpl in Ndst1f/fAlbCre+ mice would argue that altering hepatocyte HS does not affect its distribution, consistent with the idea that the enzyme is mostly bound to endothelial HS (7). Genetic studies of Lpl in mice have not been insightful with respect to this problem, since deletion of the enzyme leads to neonatal lethality (61). Thus, we cannot exclude a role for Lpl in hepatic clearance of TRLs.
HSPGs could also act directly as endocytic receptors. Liver contains transcripts for numerous membrane-associated proteoglycans, including syndecan-1, -2, and -4 and glypican-1 and -4 (J.R. Bishop, K.I. Stanford, and J.D. Esko, unpublished observations). Since membrane proteoglycans undergo clustering and constitutive endocytosis (4, 22, 62, 63), lipoproteins bound to the proteoglycans could “piggy-back” into the cell. Studies of isolated hepatocytes showed that altering HS affected binding and uptake of VLDL particles, consistent with this idea (Figure 5C). Interestingly, inactivation of Ndst1 in endothelial cells had no effect on lipoprotein metabolism, suggesting that the proteoglycans produced by the endothelium lining the space of Disse do not play a direct role in hepatic clearance.
It is also important to note that the inactivation of Ndst1 in AlbCre mice led to partial reduction in sulfation of HS, suggesting that the contribution of HS to TRL clearance may be underestimated by this approach. More profound alteration of HS structure, e.g., by mutation of the copolymerase or other sulfotransferases, might lead to greater accumulation of remnants. Furthermore, this approach could help sort out the various roles of apoE, Lpl, and HL in clearance if differences in binding to HS with respect to N-, 2-O-, and 6-O-sulfation and epimerization exist. We recently obtained a loxP-flanked allele of the uronyl 2-O-sulfotransferase (L. Wang and J.D. Esko, unpublished observations), and a conditional allele of the copolymerase Ext1 has been reported (64). The availability of these mutants and strains altered in proteoglycan core proteins should make it possible to unravel the role of HSPGs and the various apolipoproteins and lipases in lipoprotein clearance.
Can alterations in HS explain human hyperlipidemias?
The phenotype of Ndst1-deficient mice is similar to that of patients with hypertriglyceridemia associated with elevated levels of VLDL particles (65). These patients persistently exhibit elevated plasma triglyceride levels, but plasma cholesterol and phospholipid levels usually remain close to normal limits, much like in Ndst1-deficient mice (Figure 2). The etiology of hypertriglyceridemia in such patients is complex and is frequently associated with indices of metabolic syndrome. Other factors that associate with hypertriglyceridemia include chronic alcohol consumption, excessive carbohydrate intake, and certain drugs. Diabetes also results in accumulation of plasma triglycerides, which increases risk for atherosclerosis and heart disease in affected patients. Interestingly, livers from insulin-deficient rodents show reduced HS production (66, 67), reduced sulfation of the chains (68), and decreased expression of Ndst1 (69, 70). The accumulation of TRLs in Ndst1-deficient mice lends credence to the idea that one cause of triglyceride accumulation in diabetes, and possibly other disorders, might be related to aberrant expression of liver HS.
Taken together, these clinical correlations support the idea that subtle changes in liver HS could be an under lying cause of human dyslipidemias. More than 25 enzymes are responsible for HS biosynthesis, and their expression is highly regulated (3, 71). Changes in expression or activity in any of these enzymes could alter liver HS fine structure and affect lipoprotein catabolism. Based on the studies reported here, patients predisposed to mild but clinically relevant hyperlipidemias should be screened for changes in HS structure and polymorphisms in the relevant genes involved in making HS (4).
Methods
Animals.
Mice bearing Ndst1 exon 2 flanked by loxP recombination sites (Ndst1f) were described previously (43, 47). AlbCre transgenic mice and Ldlr mice were obtained from The Jackson Laboratory. All strains were fully backcrossed onto the C57BL/6J background. All animals were housed in Association for Assessment and Accreditation of Laboratory Animal Care–approved vivaria in the School of Medicine, University of California San Diego, following standards and procedures approved by the local Institutional Animal Care and Use Committee for the ethical use of animals in experiments. Mice were weaned at 3 weeks, maintained on a 12-hour light/12-hour dark cycle, and fed water and standard rodent chow ad libitum. Mice were genotyped by PCR as described in ref. 47.
AdCre delivery.
AdCre was made by the UCSD Vector Development Core Facility. AdCre (4 × 1011 particles) diluted in PBS was injected via the tail vein into 6 adult Ldlr–/–Ndst1f/fAlbCre+ mice (50). After 3 weeks, plasma samples were taken from AdCre-injected and control mice and analyzed for changes in lipoprotein content. Hepatocytes were also isolated and analyzed for Ndst1 gene recombination by Southern blotting.
Southern blot analysis.
Hepatocytes were isolated as described previously (72), and DNA was purified using a DNeasy Tissue Kit (QIAGEN). Gene inactivation was quantified by Southern blot analysis. Genomic DNA (25 μg) was digested overnight with BglII and HindIII and separated on 1% agarose before transfer overnight to nitrocellulose. The membrane was UV-crosslinked and hybridized to an Ndst1-specific probe double-labeled with [α32P]dCTP and [α32P]dATP (EasyTides; PerkinElmer). The membrane was washed and exposed to x-ray film. Films were scanned and bands were quantitated using Adobe Photoshop.
HS purification and analysis.
HS was isolated from whole livers, essentially as described previously for its isolation from other tissues (43). Livers were homogenized, collagenase digested, and treated overnight with Pronase (2 mg/ml; Roche Diagnostics) to degrade proteins, followed by purification of the glycopeptides by anion exchange chromatography using DEAE Sephacel (Amersham Biosciences). Columns were washed with low-salt buffer (0.15 M NaCl in 20 mM sodium acetate; pH 6.0) to retain low-sulfated chains and eluted with 1 M NaCl. Glycans were released from peptides by β-elimination and repurified by anion exchange chromatography. Uronic acid concentration was determined by carbazole assay (73). HS composition was determined by disaccharide analysis. Chains were digested with heparin lyases I, II, and III (Seikagaku Corp.), and the liberated disaccharides were resolved by anion exchange HPLC with post-column derivatization and fluorescence detection (74). Molar percentages were calculated based on the relative area under each peak.
Radiolabeled HS was obtained from cultured hepatocytes grown in the presence of 50 μCi/ml of 35SO4 (655 Ci/mmol; PerkinElmer) for 24 hours. Cells and conditioned medium were combined, digested as described above, β-eliminated, and treated with chondroitinase ABC to remove chondroitin [35S]sulfate. Samples of purified heparan [35S]sulfate chains (104 cpm) were incubated for 20 minutes with 10 μg of recombinant lipid-free apoE (Invitrogen) or FGF2 (Selective Genetics) in physiological saline with and without 100 μg/ml of unfractionated heparin. Bound material was collected on nitrocellulose membranes by fast vacuum filtration as described previously (75).
Lipoprotein analysis.
Total lipids were measured in blood samples drawn from overnight-fasted animals by retro-orbital sinus bleeds. Lipoproteins were prepared from blood drawn by cardiac puncture using sequential preparative ultracentrifugation according to established methods (76). Pooled plasma samples (n = 10) were centrifuged for 20 hours at 135,000 g in a Beckman 50.3Ti rotor at d = 1.019 g/ml to collect VLDL and IDL. LDL was then collected as the d = 1.019–1.065 g/ml fraction and subsequently HDL as d = 1.065–1.21 g/ml fraction. Isolated lipoproteins were dialyzed against PBS and analyzed for lipid content. Plasma lipoproteins were also separated by gel filtration FPLC. Pooled plasma samples were loaded on a Superose 12 FPLC column (Amersham Biosciences) in 0.15 M sodium chloride containing 1 mM EDTA and 0.02% sodium azide, pH 7.4, and 0.25-ml fractions were collected (0.1 ml/min). Total cholesterol and triglyceride levels were determined enzymatically using an automated reader (Cobas Mira; Roche Diagnostics) and kits: Cholesterol High-Performance Reagent (Roche Diagnostics) and Triglyceride-SL (Diagnostic Chemicals Ltd.). Protein concentrations were determined by BCA Assay (Pierce Biotechnology).
Analysis of apolipoprotein composition.
Equal volumes of the d < 1.019 g/ml lipoproteins were concentrated on Microcon-30s (Millipore), and proteins were resolved by SDS-PAGE on 4%–15% Ready Gels (Bio-Rad). Proteins were visualized by Coomassie blue or silver stain or proteins were transferred to 0.45-μm nitrocellulose (Bio-Rad). Membranes were blocked in Tris-buffered saline containing Tween-20 and 5% bovine serum albumin for 1 hour, then incubated overnight at 4°C with goat anti-mouse apoB-48/B-100 (1:2,000), apoE (1:5,000), and apoAI (1:200) polyclonal antibodies (Santa Cruz Biotechnology Inc.). Membranes were washed and incubated with HRP-conjugated donkey anti-goat secondary antibody (1:5,000) and washed and developed with ECL West Pico SuperSignal Reagents (Pierce Biotechnology).
Lipoproteins (100 μg of protein) were delipidated with chloroform/methanol (2:1, vol/vol) and digested with trypsin (0.1%) (77). Peptides were dissolved in 5% acetonitrile/0.1% formic acid and eluted stepwise from a reverse-phase C18 capillary column directly into a tandem mass spectrometer (John Yates, The Scripps Research Institute, La Jolla, California, USA). The identities of the peptides were determined by searching the tandem mass spectra against a total mouse proteome database using SEQUEST software and a computer array (78).
Quantitative PCR.
RNA was isolated from purified hepatocytes from Ndst1f/fAlbCre– and Ndst1f/fAlbCre+ mice (TRI zol Reagent), reverse transcribed (SuperScript III Reverse Transcriptase; Invitrogen) and amplified using gene-specific primers to LDLR and LRP1. cDNA from Ldlr–/–Ndst1f/fAlbCre+ mice served as a negative control, and total mouse cDNA (Ambion) served as positive control.
Quantitation was done by the 2–ΔΔCt method using GAPDH as a control RNA (79). Threshold cycle (Ct) values from triplicate assays were used to calculate fold expression according to the Stratagene manual. Results were verified in 2 independent assays.
Lipase assays.
Pre-heparin plasma (plasma levels before heparin administration) was measured after a 5-hour fast. Post-heparin plasma (plasma levels after heparin administration) was measured 15 minutes after tail vein injection of 100 U unfractionated heparin/kg body weight. HL and Lpl activities were determined using standard assays (80, 81), and lipase mass was determined using ELISA (82, 83).
Triglyceride secretion rate.
The triglyceride secretion rate was determined by the method of Hirano et al. (51). Briefly, 500 mg/kg body weight of Triton WR-1339 (Sigma-Aldrich) was injected via the tail vein into mice fasted for 5 hours, and triglyceride concentrations were measured in plasma samples taken 30, 60, and 90 minutes after injection. The secretion rate expressed in milligrams per minute was calculated from the increment in triglyceride concentration per minute multiplied by the plasma volume of the mouse (estimated as 3.5% of body weight in grams).
VLDL plasma clearance.
Human VLDL (d < 1.006 g/ml) was isolated from healthy, fasting volunteers. Mice were fasted for 5 hours and injected with 10 μg human VLDL protein via the tail vein. Serial samples were taken by retro-orbital sinus bleeds at 1, 10, and 20 minutes after injection. The amount of human VLDL remaining in the plasma was determined by sandwich ELISA, utilizing mAb MB47, specific for human apoB-100 (52). MB47 does not bind to murine apoB-100. U-bottom 96-well plates were coated overnight with MB47 at 5 μg/ml in Tris-buffered saline, followed by addition of nonsaturating amounts of plasma (typically, 1:1,000 dilution of mouse plasma) to capture human VLDL. Bound VLDL was detected using biotinylated goat anti-human apoB-100 (BioDesign International), followed by alkaline phosphatase–labeled neutrAvidin (Pierce Biotechnology). Plates were developed with Lumi-Phos 530 (Lumigen) and read in a Dynex Technologies MLX Microtiter Plate Luminometer. Half-times were calculated using linear regression to extrapolate the clearance rates.
Vitamin A fat tolerance testing.
Vitamin A fat tolerance testing was done essentially as described previously (13). Briefly, 27.0 μCi [11,12-3H]retinol (44.4 Ci/mmol; PerkinElmer) in ethanol was mixed with 1 ml of corn oil (Sigma-Aldrich). Each mouse received 200 μl by oral gavage. Blood was sampled at the times indicated by retro-orbital sinus bleed, and radioactivity was measured in triplicate by scintillation counting.
Liver/hepatocyte VLDL uptake.
Human VLDL was labeled with DiI (Invitrogen) as described previously (84). Human VLDL (1 mg) premixed with lipoprotein-deficient serum (2 ml) was incubated with 100 μl DiI (3 mg/ml in DMSO) for at least 8 hours at 37°C. DiI-VLDL was overlaid with PBS, centrifuged at 180,000 g for 8–12 hours, dialyzed against saline/EDTA, and filtered (0.8 μm). Liver uptake of DiI-VLDL was determined in LDLR-deficient mice as described previously (36, 37). DiI-VLDL (100 μg) or saline was injected by tail vein. After 20 minutes, the liver was perfused with PBS/EDTA followed by 4% paraformaldehyde. Livers were snap-frozen, and 20-μm sections were cut on a cryostat and analyzed for fluorescence by deconvolution microscopy. Images were captured with a DeltaVision Restoration microscope system (AppliedPrecision) using a Photometrics Sony Coolsnap HQ charge-coupled device (CCD) camera system attached to an inverted, wide-field fluorescence microscope (Nikon TE200). Optical sections were acquired using a 40× Nikon (NA 1.3) oil immersion objective in 0.2-μm steps in the z axis. The resolution of 40× images was enhanced using DeltaVision’s deconvolution process. DiI and DAPI fluorescence was detected using a standard DAPI, FITC, Rhodamine, and CY-5 filter set. Images were saved, processed, and analyzed on SGI workstations (O2, Octane) using the DeltaVision software package softWoRx (version 2.50). Quantification of fluorescence was accomplished using only the linear range of the digital camera.
Primary hepatocytes were isolated as described previously and grown on 22-mm coverslips in 6-well plates (85). After 24 hours, cells were serum starved in complete growth media supplemented with 0.5 mg/ml BSA for 4 hours and then incubated with 5 μg/ml of DiI-VLDL in serum-free media for 1 hour. Coverslips were then washed 3 times in PBS, fixed in 3% formalin, mounted onto slides with ProLong, and stained with DAPI (Invitrogen). Images were obtained as described above.
Statistics.
Statistical analyses were performed using Prism 4.0c (GraphPad Software). All data are expressed as mean values ± SD. Significance was determined using an unpaired Student’s (2-tailed) t test. Significance was taken as P < 0.05.
Acknowledgments
We thank David Ditto, Jennifer Pattison, Joseph Juliano, Andrew Li, Sulaba Argade, Lisa Wiggleton, Arrate Mallabiabarrena, and Elizabeth R. Miller (University of California, San Diego) for excellent technical assistance. We also acknowledge use of the UCSD Shared Resource Core Facilities in Lipid Analysis, Hematology, Histology, Glycotechnology, and Digital Imaging. We thank Joachim Herz (University of Texas Southwestern Medical Center) and Linda Curtiss (Scripps Research Institute, La Jolla) for helpful discussions and John Yates (Scripps Research Institute, La Jolla) for help with mass spectrometry. This work was supported by NIH grants GM33063 and HL57345 (to J.D. Esko), a National Science Foundation Graduate Research Fellowship (to J.M. MacArthur), and Specialized Centers of Research (SCOR) grant HL56989 (to J.L. Witztum).
Footnotes
Nonstandard abbreviations used: AdCre, adenovirus Cre; CR, chylomicron remnant; FPLC, fast-phase liquid chromatography; GlcNAc, N-acetyl- d-glucosamine; GlcNS, N-sulfo- d-glucosamine; HL, hepatic lipase; HS, heparan sulfate; HSPG, heparan sulfate proteoglycan; IDL, intermediate-density lipoprotein; LDLR, LDL receptor; Lpl, lipoprotein lipase; LRP, LDLR-related protein; Ndst1, N-deacetylase/N-sulfotransferase; TRL, triglyceride-rich lipoprotein; ΔUA, unsaturated uronic acid.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:153–164 (2007). doi:10.1172/JCI29154
Jennifer M. MacArthur’s present address is: Porter Novelli Life Sciences, Boston, Massachusetts, USA.
Lianchun Wang’s present address is: Complex Carbohydrate Research Center, University of Georgia, Athens, Georgia, USA.
Joseph R. Bishop and Jennifer M. MacArthur contributed equally to this work.
See the related Commentary beginning on page 94.
References
- 1. Conrad, H.E. 1998. Heparin-binding proteins. Academic Press. San Diego, California, USA. 527 pp. [Google Scholar]
- 2.Bernfield M., et al. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 3.Esko J.D., Selleck S.B. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 4.Williams K.J., Fuki I.V. Cell-surface heparan sulfate proteoglycans: dynamic molecules mediating ligand catabolism. Curr. Opin. Lipidol. 1997;8:253–262. doi: 10.1097/00041433-199710000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Belting M. Heparan sulfate proteoglycan as a plasma membrane carrier. Trends Biochem. Sci. 2003;28:145–151. doi: 10.1016/S0968-0004(03)00031-8. [DOI] [PubMed] [Google Scholar]
- 6.Williams K.J., et al. Mechanisms by which lipoprotein lipase alters cellular metabolism of lipoprotein(a), low density lipoprotein, and nascent lipoproteins. Roles for low density lipoprotein receptors and heparan sulfate proteoglycans. J. Biol. Chem. 1992;267:13284–13292. [PubMed] [Google Scholar]
- 7.Saxena U., Klein M.G., Goldberg I.J. Identification and characterization of the endothelial cell surface lipoprotein lipase receptor. J. Biol. Chem. 1991;266:17516–17521. [PubMed] [Google Scholar]
- 8.Mahley R.W., Ji Z.S. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J. Lipid Res. 1999;40:1–16. [PubMed] [Google Scholar]
- 9.Vilaro S., et al. Lipoprotein lipase in liver. Release by heparin and immunocytochemical localization. Biochim. Biophys. Acta. 1988;959:106–117. doi: 10.1016/0005-2760(88)90021-5. [DOI] [PubMed] [Google Scholar]
- 10.Sanan D.A., Fan J., Bensadoun A., Taylor J.M. Hepatic lipase is abundant on both hepatocyte and endothelial cell surfaces in the liver. J. Lipid Res. 1997;38:1002–1013. [PubMed] [Google Scholar]
- 11.Rubinsztein D.C., et al. Chylomicron remnant clearance from the plasma is normal in familial hypercholesterolemic homozygotes with defined receptor defects. J. Clin. Invest. 1990;86:1306–1312. doi: 10.1172/JCI114839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kita T., et al. Hepatic uptake of chylomicron remnants in WHHL rabbits: a mechanism genetically distinct from the low density lipoprotein receptor. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3623–3627. doi: 10.1073/pnas.79.11.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishibashi S., et al. Role of the low density lipoprotein (LDL) receptor pathway in the metabolism of chylomicron remnants. A quantitative study in knockout mice lacking the LDL receptor, apolipoprotein E, or both. J. Biol. Chem. 1996;271:22422–22427. doi: 10.1074/jbc.271.37.22422. [DOI] [PubMed] [Google Scholar]
- 14.Cooper A.D. Hepatic uptake of chylomicron remnants. J. Lipid Res. 1997;38:2173–2192. [PubMed] [Google Scholar]
- 15.Rohlmann A., Gotthardt M., Hammer R.E., Herz J. Inducible inactivation of hepatic LRP gene by cre-mediated recombination confirms role of LRP in clearance of chylomicron remnants. . J. Clin. Invest. 1998;101:689–695. doi: 10.1172/JCI1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujino T., et al. Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc. Natl. Acad. Sci. U. S. A. 2003;100:229–234. doi: 10.1073/pnas.0133792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji Z.S., et al. Role of heparan sulfate proteoglycans in the binding and uptake of apolipoprotein E-enriched remnant lipoproteins by cultured cells. J. Biol. Chem. 1993;268:10160–10167. [PubMed] [Google Scholar]
- 18.Ji Z.S., Fazio S., Mahley R.W. Variable heparan sulfate proteoglycan binding of apolipoprotein E variants may modulate the expression of type III hyperlipoproteinemia. J. Biol. Chem. 1994;269:13421–13428. [PubMed] [Google Scholar]
- 19.Ji Z.S., et al. Enhanced binding and uptake of remnant lipoproteins by hepatic lipase-secreting hepatoma cells in culture. J. Biol. Chem. 1994;269:13429–13436. [PubMed] [Google Scholar]
- 20.Ji Z.S., Mahley R.W. Lactoferrin binding to heparan sulfate proteoglycans and the LDL receptor-related protein. Further evidence supporting the importance of direct binding of remnant lipoproteins to HSPG. Arterioscler. Thromb. 1994;14:2025–2031. doi: 10.1161/01.atv.14.12.2025. [DOI] [PubMed] [Google Scholar]
- 21.Al-Haideri M., et al. Heparan sulfate proteoglycan-mediated uptake of apolipoprotein E-triglyceride-rich lipoprotein particles: a major pathway at physiological particle concentrations. Biochemistry. 1997;36:12766–12772. doi: 10.1021/bi9631024. [DOI] [PubMed] [Google Scholar]
- 22.Fuki I.V., et al. The syndecan family of proteoglycans. Novel receptors mediating internalization of atherogenic lipoproteins in vitro. J. Clin. Invest. 1997;100:1611–1622. doi: 10.1172/JCI119685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji Z.S., Dichek H.L., Miranda R.D., Mahley R.W. Heparan sulfate proteoglycans participate in hepatic lipase- and apolipoprotein E-mediated binding and uptake of plasma lipoproteins, including high density lipoproteins. J. Biol. Chem. 1997;272:31285–31292. doi: 10.1074/jbc.272.50.31285. [DOI] [PubMed] [Google Scholar]
- 24.Zeng B.J., Mortimer B.C., Martins I.J., Seydel U., Redgrave T.G. Chylomicron remnant uptake is regulated by the expression and function of heparan sulfate proteoglycan in hepatocytes. . J. Lipid Res. 1998;39:845–860. [PubMed] [Google Scholar]
- 25.Fuki I.V., Iozzo R.V., Williams K.J. Perlecan heparan sulfate proteoglycan. A novel receptor that mediates a distinct pathway for ligand catabolism. J. Biol. Chem. 2000;275:25742–25750. doi: 10.1074/jbc.M909173199. [DOI] [PubMed] [Google Scholar]
- 26.Wilsie L.C., Orlando R.A. The low density lipoprotein receptor-related protein complexes with cell surface heparan sulfate proteoglycans to regulate proteoglycan-mediated lipoprotein catabolism. J. Biol. Chem. 2003;278:15758–15764. doi: 10.1074/jbc.M208786200. [DOI] [PubMed] [Google Scholar]
- 27.Libeu C.P., et al. New insights into the heparan sulfate proteoglycan-binding activity of apolipoprotein E. J. Biol. Chem. 2001;276:39138–39144. doi: 10.1074/jbc.M104746200. [DOI] [PubMed] [Google Scholar]
- 28.Dong J., et al. Interaction of the N-terminal domain of apolipoprotein E4 with heparin. Biochemistry. 2001;40:2826–2834. doi: 10.1021/bi002417n. [DOI] [PubMed] [Google Scholar]
- 29.Spillmann D., Lookene A., Olivecrona G. Isolation and characterization of low sulfated heparan sulfate sequences with affinity for lipoprotein lipase. J. Biol. Chem. 2006;281:23405–23413. doi: 10.1074/jbc.M604702200. [DOI] [PubMed] [Google Scholar]
- 30.Parthasarathy N., et al. Oligosaccharide sequences of endothelial cell surface heparan sulfate proteoglycan with affinity for lipoprotein lipase. J. Biol. Chem. 1994;269:22391–22396. [PubMed] [Google Scholar]
- 31.Sendak R.A., Berryman D.E., Gellman G., Melford K., Bensadoun A. Binding of hepatic lipase to heparin: identification of specific heparin-binding residues in two distinct positive charge clusters. J. Lipid Res. 2000;41:260–268. [PubMed] [Google Scholar]
- 32.Yu W., Hill J.S. Mapping the heparin-binding domain of human hepatic lipase. Biochem. Biophys. Res. Commun. 2006;343:659–665. doi: 10.1016/j.bbrc.2006.02.175. [DOI] [PubMed] [Google Scholar]
- 33.Lyon M., Deakin J.A., Gallagher J.T. Liver heparan sulfate structure. A novel molecular design. J. Biol. Chem. 1994;269:11208–11215. [PubMed] [Google Scholar]
- 34.Vongchan P., et al. Structural characterization of human liver heparan sulfate. Biochim. Biophys. Acta. 2005;1721:1–8. doi: 10.1016/j.bbagen.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Windler E., et al. Differences in the mechanisms of uptake and endocytosis of small and large chylomicron remnants by rat liver. Hepatology. 1996;24:344–351. doi: 10.1053/jhep.1996.v24.pm0008690403. [DOI] [PubMed] [Google Scholar]
- 36.Ji Z.S., Sanan D.A., Mahley R.W. Intravenous heparinase inhibits remnant lipoprotein clearance from the plasma and uptake by the liver: in vivo role of heparan sulfate proteoglycans. . J. Lipid Res. 1995;36:583–592. [PubMed] [Google Scholar]
- 37.Mortimer B.C., Beveridge D.J., Martins I.J., Redgrave T.G. Intracellular localization and metabolism of chylomicron remnants in the livers of low density lipoprotein receptor-deficient mice and apoE-deficient mice. Evidence for slow metabolism via an alternative apoE-dependent pathway. J. Biol. Chem. 1995;270:28767–28776. doi: 10.1074/jbc.270.48.28767. [DOI] [PubMed] [Google Scholar]
- 38.Mann W.A., et al. Dominant expression of type III hyperlipoproteinemia. Pathophysiological insights derived from the structural and kinetic characteristics of ApoE-1 (Lys146-->Glu). J. Clin. Invest. 1995;96:1100–1107. doi: 10.1172/JCI118096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mann W.A., et al. Apolipoprotein E isoforms and rare mutations: parallel reduction in binding to cells and to heparin reflects severity of associated type III hyperlipoproteinemia. J. Lipid Res. 1995;36:517–525. [PubMed] [Google Scholar]
- 40.Lutz E.P., et al. Heparin-binding defective lipoprotein lipase is unstable and causes abnormalities in lipid delivery to tissues. J. Clin. Invest. 2001;107:1183–1192. doi: 10.1172/JCI11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ringvall M., et al. Defective heparan sulfate biosynthesis and neonatal lethality in mice lacking N-deacetylase/N-sulfotransferase-1. J. Biol. Chem. 2000;275:25926–25930. doi: 10.1074/jbc.C000359200. [DOI] [PubMed] [Google Scholar]
- 42.Fan G., et al. Targeted disruption of NDST-1 gene leads to pulmonary hypoplasia and neonatal respiratory distress in mice. FEBS Lett. 2000;467:7–11. doi: 10.1016/s0014-5793(00)01111-x. [DOI] [PubMed] [Google Scholar]
- 43.Grobe K., et al. Cerebral hypoplasia and craniofacial defects in mice lacking heparan sulfate Ndst1 gene function. Development. 2005;132:3777–3786. doi: 10.1242/dev.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ledin J., et al. Heparan sulfate structure in mice with genetically modified heparan sulfate production. J. Biol. Chem. 2004;279:42732–42741. doi: 10.1074/jbc.M405382200. [DOI] [PubMed] [Google Scholar]
- 45.Forsberg E., et al. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature. 1999;400:773–776. doi: 10.1038/23488. [DOI] [PubMed] [Google Scholar]
- 46.Humphries D.E., et al. Heparin is essential for the storage of specific granule proteases in mast cells. Nature. 1999;400:769–772. doi: 10.1038/23481. [DOI] [PubMed] [Google Scholar]
- 47.Wang L., Fuster M., Sriramarao P., Esko J.D. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat. Immunol. 2005;6:902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 48.Postic C., Magnuson M.A. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 49.Ishibashi S., et al. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J. Clin. Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohlmann A., Gotthardt M., Willnow T.E., Hammer R.E., Herz J. Sustained somatic gene inactivation by viral transfer of Cre recombinase. Nat. Biotechnol. 1996;14:1562–1565. doi: 10.1038/nbt1196-1562. [DOI] [PubMed] [Google Scholar]
- 51.Williams K.J., Brocia R.W., Fisher E.A. The unstirred water layer as a site of control of apolipoprotein B secretion. J. Biol. Chem. 1990;265:16741–16744. [PubMed] [Google Scholar]
- 52.Hirano T., et al. Apoprotein C-III deficiency markedly stimulates triglyceride secretion in vivo: comparison with apoprotein E. Am. J. Physiol. Endocrinol. Metab. 2001;281:E665–E669. doi: 10.1152/ajpendo.2001.281.4.E665. [DOI] [PubMed] [Google Scholar]
- 53.Young S.G., Smith R.S., Hogle D.M., Curtiss L.K., Witztum J.L. Two new monoclonal antibody-based enzyme-linked assays of apolipoprotein B. Clin. Chem. 1986;32:1484–1490. [PubMed] [Google Scholar]
- 54.Guimond S., Maccarana M., Olwin B.B., Lindahl U., Rapraeger A.C. Activating and inhibitory heparin sequences for FGF-2 (basic FGF). Distinct requirements for FGF-1, FGF-2, and FGF-4. . J. Biol. Chem. 1993;268:23906–23914. [PubMed] [Google Scholar]
- 55.Bensadoun A., Berryman D.E. Genetics and molecular biology of hepatic lipase. Curr. Opin. Lipidol. 1996;7:77–81. doi: 10.1097/00041433-199604000-00005. [DOI] [PubMed] [Google Scholar]
- 56.Santamarina-Fojo S., Gonzalez-Navarro H., Freeman L., Wagner E., Nong Z. Hepatic lipase, lipoprotein metabolism, and atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2004;24:1750–1754. doi: 10.1161/01.ATV.0000140818.00570.2d. [DOI] [PubMed] [Google Scholar]
- 57.Otarod J.K., Goldberg I.J. Lipoprotein lipase and its role in regulation of plasma lipoproteins and cardiac risk. Curr. Atheroscler. Rep. 2004;6:335–342. doi: 10.1007/s11883-004-0043-4. [DOI] [PubMed] [Google Scholar]
- 58.Magoori K., et al. Severe hypercholesterolemia, impaired fat tolerance, and advanced atherosclerosis in mice lacking both low density lipoprotein receptor-related protein 5 and apolipoprotein E. J. Biol. Chem. 2003;278:11331–11336. doi: 10.1074/jbc.M211987200. [DOI] [PubMed] [Google Scholar]
- 59.Hamilton R.L., Wong J.S., Guo L.S., Krisans S., Havel R.J. Apolipoprotein E localization in rat hepatocytes by immunogold labeling of cryothin sections. J. Lipid Res. 1990;31:1589–1603. [PubMed] [Google Scholar]
- 60.Homanics G.E., et al. Mild dyslipidemia in mice following targeted inactivation of the hepatic lipase gene. J. Biol. Chem. 1995;270:2974–2980. doi: 10.1074/jbc.270.7.2974. [DOI] [PubMed] [Google Scholar]
- 61.Weinstock P.H., et al. Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. . J. Clin. Invest. 1995;96:2555–2568. doi: 10.1172/JCI118319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yanagishita M. Glycosylphosphatidylinositol-anchored and core protein-intercalated heparan sulfate proteoglycans in rat ovarian granulosa cells have distinct secretory, endocytotic, and intracellular degradative pathways. J. Biol. Chem. 1992;267:9505–9511. [PubMed] [Google Scholar]
- 63.Belting M. Heparan sulfate proteoglycan as a plasma membrane carrier. Trends Biochem. Sci. 2003;28:145–151. doi: 10.1016/S0968-0004(03)00031-8. [DOI] [PubMed] [Google Scholar]
- 64.Inatani M., Irie F., Plump A.S., Tessier-Lavigne M., Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- 65. Havel, R.J., and Kane, J.P. 2001. Structure and metabolism of plasma lipoproteins. In The metabolic and molecular bases of inherited disease. C.R. Scriver et al., editors. McGraw-Hill. New York, New York, USA. 2705–2716. [Google Scholar]
- 66.Ebara T., et al. Delayed catabolism of apoB-48 lipoproteins due to decreased heparan sulfate proteoglycan production in diabetic mice. J. Clin. Invest. 2000;105:1807–1818. doi: 10.1172/JCI8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olsson U., et al. Changes in matrix proteoglycans induced by insulin and fatty acids in hepatic cells may contribute to dyslipidemia of insulin resistance. Diabetes. 2001;50:2126–2132. doi: 10.2337/diabetes.50.9.2126. [DOI] [PubMed] [Google Scholar]
- 68.Kjellen L., Bielefeld D., Hook M. Reduced sulfation of liver heparan sulfate in experimentally diabetic rats. Diabetes. 1983;32:337–342. doi: 10.2337/diab.32.4.337. [DOI] [PubMed] [Google Scholar]
- 69.Unger E., Pettersson I., Eriksson U.J., Lindahl U., Kjellén L. Decreased activity of the heparan sulfate-modifying enzyme glucosaminyl N-deacetylase in hepatocytes from streptozotocin-diabetic rats. J. Biol. Chem. 1991;266:8671–8674. [PubMed] [Google Scholar]
- 70.Williams K.J., et al. Loss of heparan N-sulfotransferase in diabetic liver: role of angiotensin II. Diabetes. 2005;54:1116–1122. doi: 10.2337/diabetes.54.4.1116. [DOI] [PubMed] [Google Scholar]
- 71.Grobe K., Esko J.D. Regulated translation of heparan sulfate N-acetylglucosamine N-deacetylase/N-sulfotransferase isozymes by structured 5′-untranslated regions and internal ribosome entry sites. J. Biol. Chem. 2002;277:30699–30706. doi: 10.1074/jbc.M111904200. [DOI] [PubMed] [Google Scholar]
- 72.Horton J.D., Shimano H., Hamilton R.L., Brown M.S., Goldstein J.L. Disruption of LDL receptor gene in transgenic SREBP-1a mice unmasks hyperlipidemia resulting from production of lipid-rich VLDL. J. Clin. Invest. 1999;103:1067–1076. doi: 10.1172/JCI6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bitter T., Muir H.M. A modified uronic acid carbazole reaction. Anal. Biochem. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- 74.Toyoda H., Nagashima T., Hirata R., Toida T., Imanari T. Sensitive high-performance liquid chromatographic method with detection for the determination of heparin and heparan sulfate in biological samples: application to human urinary heparan sulfate. J. Chromatogr. B. Biomed. Sci. Appl. . 1997;704:19–24. doi: 10.1016/s0378-4347(97)00478-7. [DOI] [PubMed] [Google Scholar]
- 75.Kreuger J., Lindahl U., Jemth P. Nitrocellulose filter binding to assess binding of glycosaminoglycans to proteins. Methods Enzymol. 2003;363:327–339. doi: 10.1016/S0076-6879(03)01062-0. [DOI] [PubMed] [Google Scholar]
- 76.Kelley J.L., Kruski A.W. Density gradient ultracentrifugation of serum lipoproteins in a swinging bucket rotor. Methods Enzymol. 1986;128:170–181. doi: 10.1016/0076-6879(86)28067-2. [DOI] [PubMed] [Google Scholar]
- 77.Rall S.C., Weisgraber K.H., Mahley R.W. Isolation and characterization of apolipoprotein E. Methods Enzymol. 1986;128:273–287. doi: 10.1016/0076-6879(86)28073-8. [DOI] [PubMed] [Google Scholar]
- 78.Wolters D.A., Washburn M.P., Yates J.R. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 79.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 80. Ehnholm, C., and Kuusi, T. 1986. Preparation, characterization, and measurement of hepatic lipase. In Methods in enzymology. J.J. Albers and J.P. Segrest, editors. Academic Press. Orlando, Florida, USA. 716–738. [DOI] [PubMed] [Google Scholar]
- 81. Iverius, P., and Ostlund-Lindqvist, A. 1986. Preparation, characterization, and measurement of lipoprotein lipase. In Methods in enzymology. J.J. Albers and J.P. Segrest, editors. Academic Press. Orlando, Florida, USA. 691–704. [DOI] [PubMed] [Google Scholar]
- 82.Cisar L.A., Bensadoun A. Enzyme-linked immunosorbent assay for rat hepatic triglyceride lipase. J. Lipid Res. 1985;26:380–386. [PubMed] [Google Scholar]
- 83.Cupp M., Bensadoun A., Melford K. Heparin decreases the degradation rate of lipoprotein lipase in adipocytes. J. Biol. Chem. 1987;262:6383–6388. [PubMed] [Google Scholar]
- 84.Pitas R.E., Innerarity T.L., Weinstein J.N., Mahley R.W. Acetoacetylated lipoproteins used to distinguish fibroblasts from macrophages in vitro by fluorescence microscopy. Arteriosclerosis. 1981;1:177–185. doi: 10.1161/01.atv.1.3.177. [DOI] [PubMed] [Google Scholar]
- 85.Bishop J.R., Crawford B.E., Esko J.D. Cell surface heparan sulfate promotes replication of Toxoplasma gondii. Infect. Immun. 2005;73:5395–5401. doi: 10.1128/IAI.73.9.5395-5401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Havel R.J., Hamilton R.L. Hepatic catabolism of remnant lipoproteins: where the action is. Arterioscler. Thromb. Vasc. Biol. 2004;24:213–215.. doi: 10.1161/01.ATV.0000115382.53810.24. [DOI] [PubMed] [Google Scholar]