Abstract
Homology search is one of the most ubiquitous bioinformatic tasks, yet it is unknown how effective the currently available tools are for identifying noncoding RNAs (ncRNAs). In this work, we use reliable ncRNA data sets to assess the effectiveness of methods such as BLAST, FASTA, HMMer, and Infernal. Surprisingly, the most popular homology search methods are often the least accurate. As a result, many studies have used inappropriate tools for their analyses. On the basis of our results, we suggest homology search strategies using the currently available tools and some directions for future development.
Compared with the relatively trivial task of protein homology search, ncRNA homology search is more challenging because of the fact that intra- and intermolecular base pairs are, in evolutionary terms, preserved to a higher degree than the sequence. The wobble GU and other noncanonical base pairs allow RNA sequences to evolve seemingly unrelated sequences along nearly neutral paths through structure space (e.g., A · U ↔ G · U ↔ G · C). Thus, specialized homology search techniques, such as nucleotide specific scoring schemes (States et al. 1991), profile hidden Markov model (profile HMMs) (Haussler et al. 1993; Krogh et al. 1994), and covariance models (CMs) (Eddy and Durbin 1994), are necessary for accurate ncRNA homology search.
The goal of this study is to identify programs that balance sensitivity (true predictions) and specificity (false predictions) for practical ncRNA homology search situations. We use large high-quality ncRNA data sets and randomized control data sets to test the 12 homology search programs summarized in Table 1. Briefly, sequences are sampled from each ncRNA data set and then used as input sequences for each algorithm against the original (true homologs) and randomized data sets. Our test data sets are composed of a subclass of ncRNAs that tend to be highly structured, and therefore there is more information for homology detection than for unconstrained ncRNAs. The algorithms that do not perform well on these data sets are not likely to perform better on more challenging classes of ncRNAs. To ensure our results reflect practical scenarios, we have used both predicted alignments and secondary structures to generate input data for the alignment and structure-based methods.
Table 1.
Program descriptions, URLs, and references for each of the 12 programs used in this study
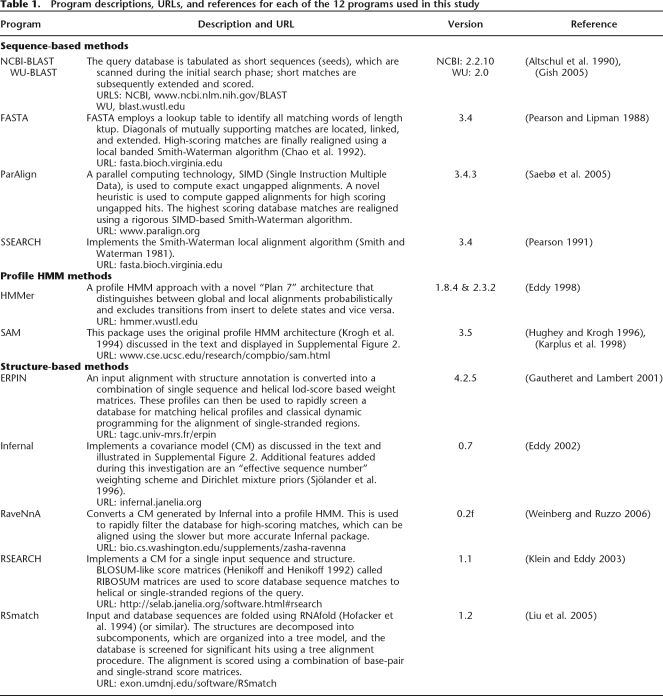
Homology search programs fall into one of three classes: sequence based methods, profile HMM methods, and structure enhanced methods (Fig. 1). In addition to evaluating homology search programs, we extend the use of ancestral sequence reconstructions (ASR) and introduce the novel phylogeny-based predictive sequence reconstruction (PSR) method for use in homology searches (Collins et al. 2003; McCormack 2003; Qian and Goldstein 2003; Cai et al. 2004) to the RNA homology search problem (see Supplemental Fig. 1). Briefly, we discuss each of these in turn.
Figure 1.
An overview of homology search methods. A Venn diagram illustrating an overview of the methods used in this study. Different methods are classified as heuristic, single sequence, profile HMM, stochastic context-free grammar (SCFG), and/or RNA specific.
The most popular homology search methods are sequence based. The local matching of two sequences has been solved by Smith and Waterman (1981) in a mathematically optimal fashion using a dynamic programming procedure. However, this method is too slow for most practical homology search situations, where the database length is large. Hence, heuristic methods such as BLAST and FASTA, which speed the search procedure but at a cost to accuracy, are often used.
Profile HMMs have been used for detecting patterns in multiple sequences (Haussler et al. 1993; Krogh et al. 1994); assessments of profile HMMs on protein data sets have proven that these are more accurate than sequence methods alone (Brenner et al. 1998; Park et al. 1998; Lindahl and Elofsson 2000; Madera and Gough 2002). The basic usage of a profile HMM is to convert an input alignment into a probabilistic model, which is used to scan a database for homologous sequences. The fundamental concept of profile HMMs can be understood by considering nucleotide frequencies in each column of an alignment. In the absence of gaps, the probability that a given sequence is generated by the same evolutionary processes as those in the alignment can be estimated by the product of position specific nucleotide frequencies. The architecture proposed by Krogh et al. (1994) (see Supplemental Fig. 2) allows for insertions and deletions in the model, and, in addition, deletions can be modeled in a position-dependent manner. To account for overrepresented sequences in the input alignment, tree-weighting schemes can be used (Durbin et al. 1998), and there are schemes to avoiding over-fitting and to account for unobserved data in the input (Sjölander et al. 1996).
Structure-enhanced methods are frequently based on CMs, which are an analog of profile HMMs that include pairwise interactions due to RNA secondary structure. Whereas profile HMMs consist of a linear HMM architecture suitable for modeling linear protein sequences, tree-like CMs model tree-like RNA secondary structures that allow for base-pairing interactions. States within the CM capture paired and unpaired regions while allowing insertions and deletions. To picture this, imagine the profile HMM model in Supplemental Figure 2 with base pairs between distant sites. Several new states need to be added to the model to accommodate this more complex structure. In the paired sites, deletions now include either a single 5′ or 3′ base or the entire base pair, and insertions can now be between either the 5′ or 3′ ends of a base pair. Bifurcation states are also included in the CM to allow for multiloops. The basic CM search procedure is analogous to the use of profile HMMs. An alignment replete with a structure annotation is provided by the user; this is used to train a CM that is specific to the input data, which can then be used to search a query database (Eddy and Durbin 1994; Durbin et al. 1998).
Nearly all of the current homology search algorithms, with the exception of some profile HMM tree-weighting schemes (Durbin et al. 1998), ignore important evolutionary information contained in the underlying phylogenetic relationships among the input sequences. For example, the branching order and distance between sequences is largely ignored. To address these shortcomings and aid the phylogenetically naive algorithms, we employ two probabilistic phylogenetic approaches: ASR and PSR. Briefly, each of these methods sample high-probability ancestral sequences that are added to the query sequences. In this way, additional information derived from the phylogenetic relationships is added to the search, in theory boosting remote homolog detection.
Caveats to algorithm assessments
There are several limitations to any algorithm assessment. We outline the most important issues below.
Test data sets
We take for granted the accuracy of structural alignments taken from the literature, many of which have been constructed using the programs we are studying. However, given this limitation, the analysis of a large and diverse data set should outweigh any possible errors due to data set inaccuracies. We make one final point regarding the nature of our test data sets; the data sets used here are conserved “ideal” ncRNAs and may not be representative of other ncRNAs. Although it is likely that other families of ncRNAs will be less conserved and less “ideal,” it seems clear that if a method fails under “ideal” circumstances, its performance is unlikely to improve in more challenging circumstances.
Tool abuse
Frequently, researchers may apply a tool to a task for which it is not designed. For example, in this study we have applied sequence-based tools to structured ncRNAs, assuming that sites are independent. This is a common but poor assumption.
Tools improve
Many of the tools tested here are recent developments and are still under active development. Hence, not all observations will remain reproducible. In fact, we hope this study helps improve future performance.
Parameter settings
The performance of some of the programs may benefit from optimizing program parameters. Here, we have attempted to capture the essential features of each algorithm by using as many parameter combinations as was practical.
During the course of this investigation, we contacted the authors of each of the programs included in this study (see Acknowledgments). We provided access to the data sets, scripts, and a preprint of the article for the authors from the BRaliBase Web site (www.binf.ku.dk/~pgardner/bralibase). We found the comments we received invaluable for minimizing the costs of the above caveats.
Results
The following discussion contains a detailed summary of the results presented in Figures 2, 3, 4 and the Supplemental Tables 1–4 and Supplemental Figures 5–7. We begin by outlining the results for each method with the parameter settings that had the optimal ranking. Unless stated otherwise, we focus on the results for the smaller query subsets with just 5 sequences because the results do not differ significantly from the analysis using larger query subsets (20 sequences). Secondly, we outline the results for our secondary tests, which include a comparison of the sequence-based methods (NCBI-BLAST, WU-BLAST, FASTA, ParAlign, and SSEARCH) with identical scoring parameters. These scoring parameters are optimized for sequence identities ranging from 65%–100% (States et al. 1991) and are referred to as the 65% scoring scheme throughout this section. The other secondary tests we present are of RNA-centric scoring schemes for sequence-based methods, the application of phylogenetic sequence reconstruction to homology searching, and a scan of a section of the human genome.
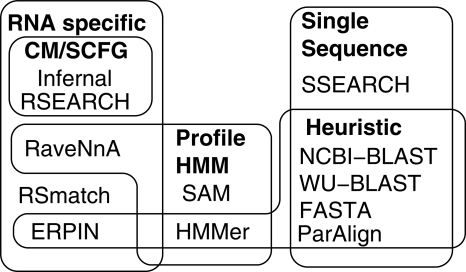
Figure 2.
A comparison of the accuracy and efficiencies of homology search methods showing only the highest-ranking parameter settings for each algorithm from Supplemental Table 1. These were NCBI-BLAST (W7, 65%), WU-BLAST (W3), FASTA, ParAlign (65%), SSEARCH, HMMer (2.3.2, local), SAM (3.5, local), ERPIN, Infernal (0.7, local), RaveNnA, RSEARCH, and RSmatch. (A,B) Boxplots of algorithm ranks for the 5 and 20 sequence subsets, respectively. The blue curves show the median sensitivity, the green curve the median specificity, and the red curve the median MCC for each of the 12 programs. These accuracy values were computed by sampling either 5 or 20 sequences from the reference databases; these were used as input(s) to each algorithm for screening both the reference and a shuffled database. (C) Boxplots of algorithm speeds in nucleotides per second. The red curve shows median initialization times for the different programs. The single sequence, profile HMM, and RNA methods are displayed in unshaded, dark shaded, and lightly shaded boxes, respectively.
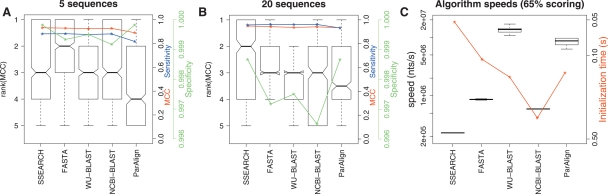
Figure 3.
A comparison of the accuracy of sequence-based methods with the 65% scoring scheme and identical scoring parameters. These boxplots show the distributions of the ranks on MCC and timing data for each of the homology search methods when using a scoring scheme optimized for nucleotide sequences with 65% identity (match = +5, mismatch = −4, gapopen = 10, gapextension = 10). (A,B) Boxplots of algorithm ranks for the 5 and 20 sequence subsets, respectively. The blue curves show the median sensitivity, the green curve the median specificity, and the red curve the median MCC for each of the 12 programs. (C) Boxplots of algorithm speeds in nucleotides per second. The red curve shows median initialization times for the different programs.
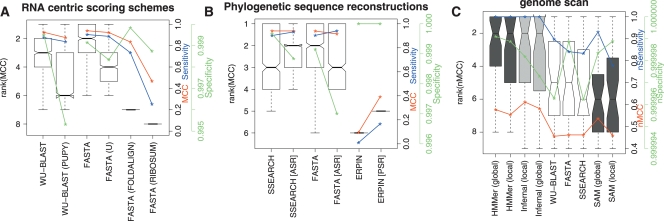
Figure 4.
A comparison of the accuracy of methods using RNA-centric scoring matrices, phylogenetic sequence reconstructions, and the genome scan results. (A) A comparison of the accuracy of sequence-based methods with score matrices optimized for ncRNA. These boxplots show the distributions of the ranks on MCC for each of the homology search methods when using one of WU-BLAST (W7), WU-BLAST (W7, PUPY), FASTA, FASTA (U), FASTA (RIBOSUM), or FASTA (FOLDALIGN). These matrices are discussed in more detail in the text. (B) A comparison of FASTA, SSEARCH, and ERPIN with and without phylogenetic sequence reconstructions included in the input. Ancestral sequence reconstruction was used in the case of FASTA and SSEARCH and posterior predictive sequences in the case of ERPIN. Both A and B show results using 5 query sequences. (C) A set of representative programs from each category were run on human chromosome 12 (coordinates 90,000,000–130,000,000; ver NCBI35). The boxplot displays algorithm ranks; additionally, median nMCC, median nSensitivity, and median nSpecificity for each algorithm are displayed using the y-axis on the right. The single sequence, profile HMM, and RNA methods are displayed in unshaded, dark shaded, and lightly shaded boxes, respectively.
Sequence-based searches
The accuracies of WU-BLAST and NCBI-BLAST were unsurprisingly similar when similar parameters were used, yet WU-BLAST was significantly faster (Fig. 3). The default scoring scheme used for NCBI-BLAST is tailored for sequences with 99% sequence homology, whereas WU-BLAST defaults are tailored for sequences with 65% sequence homology (States et al. 1991), which is more appropriate for our diverse ncRNA data sets (see Supplemental Tables 1 and 2). WU-BLAST has a more diverse array of options, including allowing a minimum seed length of 3 (W3) (compared with 7 [W7] for NCBI-BLAST). Hence, the parameter settings producing the best accuracy for WU-BLAST are not implemented in NCBI-BLAST. However, shorter seed lengths did come at a significant cost to program speed.
A comparison of FASTA and WU-BLAST (W3) is complicated given that the median ranking of WU-BLAST (W3) was higher than that of FASTA for the 5 sequence input data, but the lower quartile of the WU-BLAST (W3) ranks was much lower than that of FASTA (Fig. 3). Hence, most of the time WU-BLAST (W3) outperformed FASTA (the rankings reversed on the 20 sequence data sets). Yet, FASTA was significantly faster than WU-BLAST (W3) and compared well with NCBI-BLAST (W7, 65%) in terms of speed.
ParAlign was the fastest of the homology search tools in this study. However, ParAlign had low sensitivity compared with both FASTA and BLAST; this was true also for the 65% scoring test (see Methods and Fig. 3 for results).
SSEARCH generally outperformed the other sequence-based methods in terms of accuracy. However, SSEARCH performance was very closely correlated with WU-BLAST (W3), but WU-BLAST (W3) was significantly slower (see Supplemental Fig. 9). This observation was surprising given that SSEARCH employs no heuristics to improve speed whereas WU-BLAST (W3) demands seeds matching of at least three consecutive nucleotide positions; one would have expected the opposite to the results presented here.
Profile HMMs
The profile HMM programs evaluated here, SAM and HMMer, always outperformed the sequence-based methods. SAM usually outperformed HMMer in terms of accuracy, yet HMMer was significantly faster (Fig. 2). The results for HMMer show that version 2.3.2 is slightly better than v.1.8.4; this is in contrast to the HMMer documentation, which suggests the opposite for nucleotide sequences (because of protein-specific optimization). Earlier results based on protein data sets (Madera and Gough 2002) showed that profile searches could be improved by using SAM models and HMMer searches. We observed no such improvement on our ncRNA data sets (see Supplemental Tables 1 and 2).
Structure-enhanced homology search
The CM-based methods Infernal and RSEARCH both performed extremely well on these ncRNA data sets, providing predictions with very high sensitivity and specificity. These methods generally ranked either first or second in terms of the Matthews correlations coefficient (MCC) (see Methods for a definition) for every search. However, there was a significant cost in terms of CPU: Both take ∼1 sec to search a kilobase using a 900-MHz processor. This is about 2 orders of magnitude slower than the profile HMM and sequence-based methods.
The Infernal package was upgraded during the course of this study to version 0.7. Sean Eddy and collaborators added Dirichlet mixtures (Sjölander et al. 1996) and effective sequence number scalings to the algorithm, which resulted in a significant performance boost for both the 5 and 20 sequence data sets (see Supplemental Tables 1 and 2).
ERPIN predictions are generally very conservative, especially for the small data set or when sequence identity is high (frequently only the input data set was recovered), resulting in high-specificity yet low-sensitivity predictions. However, the speed of ERPIN was comparable with that of the sequence-based methods.
The results for RaveNnA were also good, with the algorithm ranking third after Infernal and RSEARCH in terms of accuracy. The accuracy of RaveNnA when compared with the other profile HMM methods, HMMer and SAM, was excellent. The speed of RaveNnA was about the same magnitude as SAM, which is in good agreement with theory. However, RaveNnA requires a significant initialization time (∼25 min, see Supplemental Fig. 5) from the overhead for calibrating the HMM to determining an appropriate threshold; therefore, it is only economical to use RaveNnA on larger databases.
The speed of RSmatch was nearly an order of magnitude greater than that of the structure-enhanced methods Infernal, RaveNnA, and RSEARCH; however, the accuracy was much lower.
Five versus twenty input sequences
Overall, the results were rather constant between using 5 or 20 input sequences. RSEARCH and Infernal exchanged first place; Infernal explicitly uses covariation information and hence is likely to be more powerful with larger input data sets. The performance of WU-BLAST dropped relative to the other programs.
The CM and profile HMM methods benefit from models derived from more sequences, resulting in improved specificity. The single-sequence methods, however, suffer from problems due to multiple testing, resulting in improved sensitivity at a cost to specificity.
65% scoring scheme
This study showed that the sequence-based methods perform rather similarly when using comparable parameter settings (Fig. 3; the results labeled “65%” in Supplemental Table 3; Supplemental Fig. 6). The nonheuristic method, SSEARCH, outranked the other methods in all cases. This was followed by FASTA. The two incarnations of BLAST performed almost identically; however, WU-BLAST was significantly faster than NCBI-BLAST.
RNA-centric scoring schemes
Each of the RNA-centric scoring schemes mentioned in the Methods section was given a trial (Fig. 4; Supplemental Table 4; Supplemental Fig. 7). The scoring schemes we tested are the PUPY matrix that ships with WU-BLAST, the “-U” option for FASTA, and the single-sequence components of the score matrices used by RSEARCH (Klein and Eddy 2003) and FoldAlign (Havgaard et al. 2005). These results were generally disappointing: None of the methods showed any improvement over less-specific schemes when the RNA-centric scores were used. In the case of the FoldAlign and RSEARCH score matrices, this is justified as these matrices were built specifically for structural methods rather than the sequence-based methods we have used here. We also tested a transition/transversion scoring scheme optimized for 65% sequence identity (States et al. 1991) (data not shown); the results of this test were also disappointing. This indicates that a great deal more work is required before such scoring schemes can be used for practical RNA homology search.
Application of phylogenetic sequence reconstruction to homology search
In general, the inclusion of ancestral information did not increase the performance of the more advanced methods. However, a number of methods did benefit from this approach. First, a significant improvement in the performance of ERPIN was observed for both the ASR and PSR approaches; the median sensitivity improved by a factor of 17 when PSR sequences were included in the search. One difficulty with this approach is the use of a prior on the length of the branch leading to the reconstructed sequence. We found the best results with very long prior branch lengths (≈20 expected substitutions per site). In addition to improving ERPIN, we found that the inclusion of ASRs improved the sensitivities of the sequence-based methods. This improvement, unfortunately, comes at a cost to specificity, but in the cases of FASTA and particularly SSEARCH, an overall increase in accuracy was observed (Fig. 4). Our approach for including phylogenetic information did not improve the accuracy of the profile HMM or Infernal searches where tree-weighting schemes and Dirichlet priors are used. Since these search algorithms already incorporate information similar to that obtained from the ancestral sequences, a failure to see an increase in these methods is perhaps not surprising. Our results do suggest that future work may benefit from incorporating directly phylogenetic information along with the use of improved models of RNA sequence evolution. However, further work will be required to determine whether improvements in accuracy are due to additional information from the phylogeny or to noise injection (Krogh et al. 1994).
Genome scan
To test a representative set of algorithms in a realistic usage scenario, we scanned a 40-Mb section of the human genome. This test had the additional advantage of providing a more accurate estimate of algorithm specificity. The results of the genome scan were in general agreement with the earlier results. This was a much harder test than those presented earlier. This was compounded by the fact that the genome annotation we rely on may not be completely accurate. However, HMMer performed surprisingly well on this test compared with both Infernal and SAM. Infernal had the highest median MCC, yet had a slightly lower specificity than HMMer. This behavior may be due to unannotated homologs residing within the genomic region, which would cause false false-positives for the more sensitive methods such as Infernal and SAM. Of the single-sequence methods, both WU-BLAST and FASTA performed well. WU-BLAST had a higher sensitivity but a lower specificity (Fig. 4; Supplemental Fig. 8).
Discussion
The most popular homology search methods did not necessarily perform the best in our study. These programs are optimized for rapid database searches with few false positives (high specificity), which is not always what the user requires. As a consequence many estimates of the amount of conserved DNA (Hillier et al. 2004) and number of conserved ncRNAs (Washietl et al. 2005; Pedersen et al. 2006) and nonconserved ncRNAs (Pang et al. 2006) are based on suboptimal homology search tools and hence likely to be inaccurate.
One of the most important issues for homology searching is to develop scoring schemes that discriminate signal from noise. It is clear that the popular single sequence methods do not do this, although some modest improvements may be possible. The scorings implemented in the RNA-specific probabilistic methods Infernal, RSEARCH, and RaveNnA, however, do a good job of discriminating signal from noise. The Infernal CM method was surprisingly robust to the predicted (and therefore potentially inaccurate) input alignments and secondary structures. A comparison of Infernal with predicted and reference input data shows that the only time the predictions caused a drop in performance was when the input sequences were highly similar and the secondary structure prediction was poor. This meant that there was limited covariation information from the alignment for either the secondary structure prediction tool (RNAalifold) or the covariance model to use (see Supplemental Fig. 10).
As researchers are usually likely to favor speed over accuracy, it is necessary that FASTA and BLAST have accurate scoring schemes available that such researchers can utilize. For this, RNA-optimized PAM (Dayhoff et al. 1978) and/or BLOSUM (Henikoff and Henikoff 1992) style score matrices are needed. There are sufficient data for computing these matrices from freely available sources such as the Rfam (Griffiths-Jones et al. 2003) and the rRNA databases (Cannone et al. 2002; Wuyts et al. 2002) and the statistical methods for estimating these are well established. Additionally, given that base-pair stacking is important for RNA structure, this signal may prove useful for RNA homology search and could be exploited by incorporating a dinucleotide scoring scheme into the alignment procedure (Lunter and Hein 2004).
There are few heuristics at present for rapid profile HMM and CM-based homology searches. One could, for example, apply the BLAST concept of a seed match to profile HMMs and CMs. A database could be rapidly scanned for short, ungapped high-scoring matches to the model, which could then be extended using the full profile HMM architecture, this should result in significant gains in speed at moderate costs to sensitivity.
The specificity of homology search algorithms is an important issue when scanning large amounts of (genomic) data. If the specificity is not extremely high when scanning large databases the relatively small number of true homologs may be lost in a flood of false positives. The relatively small amounts of shuffled data we were forced to use in this study (because of the glacial speeds of some of the test algorithms) meant that we did not get an accurate measure of this value. The genome scan test we ran was meant to alleviate this problem; however, it is likely that unannotated homologs also affected the determination of algorithm specificity.
The use of phylogenetic information to enhance homology search of ncRNAs on the surface seems a bit dissatisfying. Our results do clearly indicate that there is valuable information in the phylogeny that should not be ignored as a number of the methods did benefit from our simple approach. We believe this suggests that future method development will benefit from considering the phylogeny when multiple sequences are available. For example, the use of mutational maps along the phylogeny (Nielsen 2002) could be used to create a stochastic profile for profile HMM methods (Durbin et al. 1998). Alternatively, when scanning newly sequenced genomes we often know the phylogenetic relationships between the search sequences and the query genome and may also have information on how divergent they are. Using this information, one could use the PSR approach described here to add information to the search sequences without relying on arbitrary priors about the process of evolution. Our results do hold promise for those researchers who want the set of putative homologs to include all true homologs at the cost of including a larger number of false positives.
The RSmatch algorithm relies on MFE structure predictions on a single sequence, which are known to be frequently inaccurate (Gardner and Giegerich 2004). If the structure prediction phase for both the database and input sequences were based on comparative predictions, such as RNAalifold (Hofacker et al. 2002), the accuracy of this approach is likely to improve. In addition, RSmatch could be used to cluster genome-wide structure-based ncRNA predictions (Washietl et al. 2005; Pedersen et al. 2006).
Practical recommendations
On the basis of our assessment of the currently available homology search tools, we recommend a scheme for one or more input sequences that uses iterative rounds of the rapid sequence-based methods, such as WU-BLAST, FASTA, or SSEARCH, with sensible scoring schemes and a high threshold to build a training data set. These results can then be used to train CM models for Infernal searches to obtain more divergent sequences from within the lower-scoring sequence-based matches or, if possible, the original database. Profile HMMs, particularly RaveNnA, could be used instead when CMs are not practical, for example, when the sequence length is greater than 200 nucleotides or the database is large.
Throughout this work, we have focused almost entirely on method accuracy; however, of frequent concern is the computation time for a search. For example, on the basis of our timings with tRNA queries, Infernal would take ∼96 d to search the human genome on a single processor, RaveNnA would take 40 h, HMMer would take 9 h, SSEARCH would take 4 h, and WU-BLAST (W7) just 4 min. Given the ready access many groups have to computing clusters, it is reasonable to expect the more accurate methods to become more popular in the future.
Many of the currently available tools for ncRNA homology search tools are not yet performing as well as one would hope based on the results we have presented. Improvements in terms of accuracy and speed are needed. This is extremely important given the explosion of interest in ncRNAs generated by recently discovered ncRNAs such as miRNAs. Additionally, a current theory suggests that much of the apparent organism complexity not accounted for by corresponding expansions in the proteome can be attributed to regulation from the ncRNAome (Mattick 2001).
This study has implications for evolutionary studies that rely on programs tuned for high-similarity sequences. First, since the most popular programs are biased toward identifying only highly conserved homologs, the diversity of particular ncRNAs will be underestimated perhaps more severely than previously thought. Second, and particularly irksome, is that many of the most interesting homologs will be those that are or have experienced strong positive directional or diversifying selection, causing them to have diverged beyond the detection limits of the search algorithms, and will fail to be identified. By establishing how the currently available methods perform, we can gather more high-quality databases that will allow further development of our understanding as to how different families of ncRNAs evolve. With new models in hand, we can improve the search algorithms increasing the discovery of interesting homologs, otherwise unidentified, and gain better estimates of the diversity of ncRNAs across the spectrum of life.
Methods
In the following section, we outline the data sets we used for this study and the approaches we used to compare sequence-based, profile HMM, and structure-enhanced methods.
Data sets
To test homology search tools, we have obtained hand-curated databases of 602 5S rRNAs, 1114 tRNAs, and 235 U5 spliceosomal RNAs. Sequences in the databases have mean lengths of 117, 73, and 119 nucleotides, respectively (Zwieb 1997; Sprinzl et al. 1998; Szymanski et al. 2002; Griffiths-Jones et al. 2003). Nonhomologs were generated by shuffling sequences from each database to generate a new database that was 10 times larger. The shuffling process preserved dinucleotide frequencies to avoid creating artificially dissimilar sequences (Workman and Krogh 1999). Sets of 5 and 20 search sequences were sampled from the databases. These were used to scan the original and a shuffled database. A total of 583 5-sequence sets and 360 20-sequence sets were generated.
Performance measures
Three measures of performance were used to evaluate each algorithm: sensitivity, specificity, and MCC. Sensitivity measures the fraction of the positive control data set that is recovered by an algorithm and is calculated from the number of matches to the unshuffled database for both 5 and 20 sequence sets as inputs. Specificity measures the fraction of the randomized sequences that were correctly rejected when scanning the shuffled databases with the input sequence sets of 5 and 20. The third measure, MCC, combines both sensitivity and specificity to measure the overall discriminative power of each algorithm.
Thresholds
Thresholds are used to provide a cutoff for determining whether a query sequence matches the search sequence(s). Score thresholds for each algorithm are optimized based on scans of the curated and shuffled databases with a small group of query sequences that uniformly covers the different RNA families and identity ranges. Example distributions and ROC plots are illustrated in Supplemental Figures 3 and 4. Raw scores rather than e-values are used here as there are a diverse number of methods implemented for computing e-values and these are not computed at all by some methods. In this study, we are more concerned with the scores used by specific programs than the accuracy of the different e-value computations.
65% scoring scheme
To compare the sequence-based methods on an equal footing, we have included a comparison of these using parameters optimized for data sets with 65%–100% homology (States et al. 1991) (match = +5, mismatch = −4, gapopen = 10, gapextension = 10). Where a seed was required, this was made as similar as possible, W = 7 for BLAST and ktup = 6 for FASTA.
RNA-centric scoring schemes
Several of the sequence-based methods have associated scoring schemes that are designed for the unique problem of RNA homology search. Generally, these distinguish between transitions (A ↔ G and C ↔ U), which are relatively frequent during RNA evolution, and transversions (the remaining mismatch types), which are relatively infrequent. WU-BLAST has a PUPY (purine–pyrimidine) score matrix (match = +4, transition = +2, transversion = −8). By default, FASTA and SSEARCH score a +5 for matches and −4 for mismatches, yet these tools have a “−U” scoring option that tolerates G · U wobble base pairs by scoring G/A and U/C mismatches as one less than a G/G match in a strand-specific manner. In addition, RSEARCH’s RIBOSUM (Klein and Eddy 2003) and the more recent FoldAlign (Havgaard et al. 2005) score matrices use parameters estimated directly from the loop regions of large curated ncRNA alignments. We have tested these as an alternative scoring scheme for FASTA homology searches.
Genome scan
A ncRNA-enriched region of the human genome was selected for further testing of representative homology search programs from each category. We identified a 40-Mb region on chromosome 12 (coordinates 90,000,000–130,000,000; genome assembly NCBI35) that contains 5 5S rRNAs, 10 tRNAs (and 26 pseudo-tRNAs predicted by tRNAscan-SE), and 1 U5 spliceosomal RNA. We used input data sets containing ten sequences, each with a sequence identity to the associated target RNA in one of the following ranges 40%–60%, 50%–70%, 60%–80%, and 70%–85%. The pairwise sequence identities within the data sets are between 60% and 90%.
Timing
For the timing studies, two databases of 166 Mb and 332Mb, respectively, were used. Both databases contain 1114 tRNA sequences; the smaller database has one shuffled version of each of these, whereas the larger database has three. A single tRNA subset was used as a query for the timing study. The times for scanning the 2 databases are computed on (or calibrated to) a Sun Sparc v9 and 900 MHz CPU for each algorithm. From these values, the algorithm speed (nts/s) and initialization times are computed.
Phylogenetic information
The phylogenetic relationships among the search sequences include information on the branching order and their evolutionary divergence. It has been previously suggested that ancestral reconstruction of the sequences can be used to aid homology searches by supplementing the search set with reconstructed sequences (Collins et al. 2003; McCormack 2003; Qian and Goldstein 2003; Cai et al. 2004). To test whether this type of phylogenetic information will aid in identifying homologous ncRNAs, we used an empirical Bayesian approach (Huelsenbeck and Bollback 2001) to stochastically sample ancestral sequences (ASR) (details of the sampling can be found in the Supplemental Methods). The Bayesian approach has been proven (in the case of proteins) to be the most accurate method (Hall 2006). The phylogenetic tree and model parameters were estimated using MrBayes v3 (Huelsenbeck and Ronquist 2001). To accommodate the nonindependence among sites arising from secondary structure, an RNA doublet model was used to model substitutions in stem regions (Schöniger and von Haeseler 1994; Huelsenbeck and Ronquist 2001), while loop regions were modeled using the method of Hasegawa et al. (1985). In addition to reconstructing ancestral sequences at the internal nodes of the phylogeny, a novel simple method was employed to sample ancestral sequences from unobserved lineages radiating from the internal nodes of the phylogeny using a Bayesian posterior predictive (PSR) approach (Bollback 2005) (see Supplemental Methods).
Alignment and structure prediction
We used automatic structure prediction and alignment methods that previous studies have identified as being accurate for ncRNA analyses (Gardner and Giegerich 2004; Gardner et al. 2005). The alignments are computed using ProAlign (Löytynoja and Milinkovitch 2003) and consensus structures are computed from these alignments using RNAalifold (Hofacker et al. 2002).
Performance measures
Sensitivity and specificity are common measures for determining the accuracy of homology search methods.
where TP is the number of “true positives,” TN is the number of “true negatives,” FN is the number of “false negatives,” and FP is the number of “false positives.” Sensitivity measures the fraction of the positive control data set that is recovered by the program in question; the specificity measures what fraction of the randomized sequences that were correctly rejected.
A measure combining both specificity and sensitivity is useful for ranking programs. In previous studies, the
(Klein and Eddy 2003) has been used; we, however, favor the more discriminative Matthews’ correlation coefficient (MCC) as defined below:
 |
The MCC ranges from −1 for extremely inaccurate (TP = TN = 0) to 1 for very accurate predictions (FP = FN = 0).
In general, we measure TP as the number of unique sequences in the hand-curated databases that were accepted by the algorithm in question using each of the 5 or 20 input sequences. For the genome scan, TP is instead measured as the number of nucleotides that are in both a known and a predicted RNA sequence. From this it follows how TN, FP, and FN are computed in each case. To make a distinction between the regular performance measures defined here and the ones used for the genome scan, we call the latter nSensitivity, nSpecificity, and nMCC.
To ease the comparison of the different measures, we have computed the rank of each program with representative parameter settings against the other programs using MCC values for each subset of query sequences. The rank distributions are plotted in Figures 2, 3, 4.
Acknowledgments
We thank Sam Griffiths-Jones, David Ardell, Anders Krogh, Rasmus Nielsen, Zasha Weinberg, and Jeppe Vinther for useful discussions. We also thank the homology-search algorithm developers Torbjørn Rognes, Bin Tian, William R. Pearson, Robert J. Klein, Zasha Weinberg, Stephen Altschul, Daniel Gautheret, and Sean Eddy for taking the time to make useful comments on an early draft of this manuscript. Any remaining flaws are solely our responsibility. The high-performance computer clusters at UPPMAX and the University of Copenhagen Bioinformatics Centre were used to compute many of the results presented here. P.P.G. is supported by a Carlsberg Foundation Grant (21-00-0680). J.P.B. was supported by a grant from the Danish FNU.
Footnotes
[Supplemental material is available online at www.genome.org and http://www.binf.ku.dk/~pgardner/bralibase/ bralibase3/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5890907
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bollback J.P. Posterior mapping and predictive distributions. In: Nielsen R., editor. Statistical Methods in Molecular Evolution. Springer Verlag; New York: 2005. pp. 189–203. [Google Scholar]
- Brenner S., Chothia C., Hubbard T., Chothia C., Hubbard T., Hubbard T. Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl. Acad. Sci. 1998;95:6073–6078. doi: 10.1073/pnas.95.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Pei J., Grishin N.V., Pei J., Grishin N.V., Grishin N.V. Reconstruction of ancestral protein sequences and its applications. BMC Evol. Biol. 2004;4:33. doi: 10.1186/1471-2148-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone J., Subramanian S., Schnare M., Collett J., D’Souza L., Du Y., Feng B., Lin N., Madabusi L., Muller K., Subramanian S., Schnare M., Collett J., D’Souza L., Du Y., Feng B., Lin N., Madabusi L., Muller K., Schnare M., Collett J., D’Souza L., Du Y., Feng B., Lin N., Madabusi L., Muller K., Collett J., D’Souza L., Du Y., Feng B., Lin N., Madabusi L., Muller K., D’Souza L., Du Y., Feng B., Lin N., Madabusi L., Muller K., Du Y., Feng B., Lin N., Madabusi L., Muller K., Feng B., Lin N., Madabusi L., Muller K., Lin N., Madabusi L., Muller K., Madabusi L., Muller K., Muller K., et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao K.M., Pearson W.R., Miller W., Pearson W.R., Miller W., Miller W. Aligning two sequences within a specified diagonal band. Comput. Appl. Biosci. 1992;8:481–487. doi: 10.1093/bioinformatics/8.5.481. [DOI] [PubMed] [Google Scholar]
- Collins L.J., Poole A.M., Penny D., Poole A.M., Penny D., Penny D. Using ancestral sequences to uncover potential gene homologues. Appl. Bioinformatics. 2003;2:85–95. [PubMed] [Google Scholar]
- Dayhoff M., Schwartz R., Orcutt B., Schwartz R., Orcutt B., Orcutt B. Atlas of Protein Sequence and Structure. Vol. 5. National Biomedical Research Foundation; Washington, D.C: 1978. A model of evolutionary change in proteins; pp. 345–352. [Google Scholar]
- Durbin R., Eddy S.R., Krogh A., Mitchison G., Eddy S.R., Krogh A., Mitchison G., Krogh A., Mitchison G., Mitchison G. Biological sequence analysis: Probabilistic models of protein and nucleic acids. Cambridge University Press; Cambridge, UK: 1998. [Google Scholar]
- Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. A memory efficient dynamic programming algorithm for optimal structural alignment of a sequence to an RNA secondary structure. BMC Bioinformatics. 2002;3:18. doi: 10.1186/1471-2105-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R., Durbin R., Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner P.P., Giegerich R., Giegerich R. A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinformatics. 2004;5:140. doi: 10.1186/1471-2105-5-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner P.P., Wilm A., Washietl S., Wilm A., Washietl S., Washietl S. A benchmark of multiple sequence alignment programs upon structural RNAs. Nucleic Acids Res. 2005;33:2433–2439. doi: 10.1093/nar/gki541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret D., Lambert A., Lambert A. Direct RNA definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 2001;313:1003–1011. doi: 10.1006/jmbi.2001.5102. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R., Bateman A., Marshall M., Khanna A., Eddy S.R., Marshall M., Khanna A., Eddy S.R., Khanna A., Eddy S.R., Eddy S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B.G. Simple and accurate estimation of ancestral protein sequences. Proc. Natl. Acad. Sci. 2006;103:5431–5436. doi: 10.1073/pnas.0508991103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T., Kishino H., Yano T., Yano T. Dating of the human–ape splitting by molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;21:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Havgaard J.H., Lyngsø R., Stormo G.D., Gorodkin J., Lyngsø R., Stormo G.D., Gorodkin J., Stormo G.D., Gorodkin J., Gorodkin J. Pairwise local structural alignment of RNA sequences with sequence similarity less than 40% Bioinformatics. 2005;21:1815–1824. doi: 10.1093/bioinformatics/bti279. [DOI] [PubMed] [Google Scholar]
- Haussler D., Krogh A., Mian I.S., Sjölander K., Krogh A., Mian I.S., Sjölander K., Mian I.S., Sjölander K., Sjölander K. Proceedings of the Hawaii International Conference on System Sciences. IEEE Computer Society Press; Los Alimitos, CA: 1993. Protein modeling using hidden Markov models: Analysis of globins; pp. 792–802. [Google Scholar]
- Henikoff S., Henikoff J.G., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L.W., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Bork P., Burt D.W., Groenen M.A., Delany M.E., Burt D.W., Groenen M.A., Delany M.E., Groenen M.A., Delany M.E., Delany M.E., et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Hofacker I.L., Fontana W., Bonhoeffer S., Stadler P.F., Fontana W., Bonhoeffer S., Stadler P.F., Bonhoeffer S., Stadler P.F., Stadler P.F. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- Hofacker I., Fekete M., Stadler P., Fekete M., Stadler P., Stadler P. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Bollback J.P., Bollback J.P. Empirical and hierarchical Bayesian estimation of ancestral states. Syst. Biol. 2001;50:351–366. [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hughey R., Krogh A., Krogh A. Hidden Markov models for sequence analysis: Extension and analysis of the basic method. Comput. Appl. Biosci. 1996;12:95–107. doi: 10.1093/bioinformatics/12.2.95. [DOI] [PubMed] [Google Scholar]
- Karplus K., Barrett C., Hughey R., Barrett C., Hughey R., Hughey R. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- Klein R.J., Eddy S.R., Eddy S.R. RSEARCH: Finding homologs of single structured RNA sequences. BMC Bioinformatics. 2003;4:44. doi: 10.1186/1471-2105-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Brown M., Mian I.S., Sjölander K., Haussler D., Brown M., Mian I.S., Sjölander K., Haussler D., Mian I.S., Sjölander K., Haussler D., Sjölander K., Haussler D., Haussler D. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- Lindahl E., Elofsson A., Elofsson A. Identification of related proteins on family, superfamily and fold level. J. Mol. Biol. 2000;295:613–625. doi: 10.1006/jmbi.1999.3377. [DOI] [PubMed] [Google Scholar]
- Liu J., Wang J.T., Hu J., Tian B., Wang J.T., Hu J., Tian B., Hu J., Tian B., Tian B. A method for aligning RNA secondary structures and its application to RNA motif detection. BMC Bioinformatics. 2005;6:89. doi: 10.1186/1471-2105-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A., Milinkovitch M.C., Milinkovitch M.C. A hidden Markov model for progressive multiple alignment. Bioinformatics. 2003;19:1505–1513. doi: 10.1093/bioinformatics/btg193. [DOI] [PubMed] [Google Scholar]
- Lunter G., Hein J., Hein J. A nucleotide substitution model with nearest-neighbour interactions. Bioinformatics. 2004;20:I216–I223. doi: 10.1093/bioinformatics/bth901. [DOI] [PubMed] [Google Scholar]
- Madera M., Gough J., Gough J. A comparison of profile hidden Markov model procedures for remote homology detection. Nucleic Acids Res. 2002;30:4321–4328. doi: 10.1093/nar/gkf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack T.J., Comparison of K+-channel genes within the genomes of Anopheles gambiae and Drosophila melanogaster. Genome Biol. 2003;4:R58. doi: 10.1186/gb-2003-4-9-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Mapping mutations on phylogenies. Syst. Biol. 2002;51:729–732. doi: 10.1080/10635150290102393. [DOI] [PubMed] [Google Scholar]
- Pang K.C., Frith M.C., Mattick J.S., Frith M.C., Mattick J.S., Mattick J.S. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Park J., Karplus K., Barrett C., Hughey R., Haussler D., Hubbard T., Chothia C., Karplus K., Barrett C., Hughey R., Haussler D., Hubbard T., Chothia C., Barrett C., Hughey R., Haussler D., Hubbard T., Chothia C., Hughey R., Haussler D., Hubbard T., Chothia C., Haussler D., Hubbard T., Chothia C., Hubbard T., Chothia C., Chothia C. Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol. 1998;284:1201–1210. doi: 10.1006/jmbi.1998.2221. [DOI] [PubMed] [Google Scholar]
- Pearson W.R. Searching protein sequence libraries: Comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics. 1991;11:635–650. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- Pearson W.R., Lipman D.J., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen J.S., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Lander E.S., Kent J., Miller W., Haussler D., Kent J., Miller W., Haussler D., Miller W., Haussler D., Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:251–262. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B., Goldstein R.A., Goldstein R.A. Detecting distant homologs using phylogenetic tree-based hmms. Proteins. 2003;52:446–453. doi: 10.1002/prot.10373. [DOI] [PubMed] [Google Scholar]
- Saebø P.E., Andersen S.M., Myrseth J., Laerdahl J.K., Rognes T., Andersen S.M., Myrseth J., Laerdahl J.K., Rognes T., Myrseth J., Laerdahl J.K., Rognes T., Laerdahl J.K., Rognes T., Rognes T. PARALIGN: Rapid and sensitive sequence similarity searches powered by parallel computing technology. Nucleic Acids Res. 2005;33:535–539. doi: 10.1093/nar/gki423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöniger M., von Haeseler A., von Haeseler A. A stochastic model for the evolution of autocorrelated DNA sequences. Mol. Phylogenet. Evol. 1994;3:240–247. doi: 10.1006/mpev.1994.1026. [DOI] [PubMed] [Google Scholar]
- Sjölander K., Karplus K., Brown M., Hughey R., Krogh A., Mian I.S., Haussler D., Karplus K., Brown M., Hughey R., Krogh A., Mian I.S., Haussler D., Brown M., Hughey R., Krogh A., Mian I.S., Haussler D., Hughey R., Krogh A., Mian I.S., Haussler D., Krogh A., Mian I.S., Haussler D., Mian I.S., Haussler D., Haussler D. Dirich-let mixtures: A method for improved detection of weak but significant protein sequence homology. Comput. Appl. Biosci. 1996;12:327–345. doi: 10.1093/bioinformatics/12.4.327. [DOI] [PubMed] [Google Scholar]
- Smith T., Waterman M., Waterman M. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Horn C., Brown M., Ioudovitch A., Steinberg S., Horn C., Brown M., Ioudovitch A., Steinberg S., Brown M., Ioudovitch A., Steinberg S., Ioudovitch A., Steinberg S., Steinberg S. Compilation of trna sequences and sequences of trna genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- States D.J., Gish W., Altschul S.F., Gish W., Altschul S.F., Altschul S.F. Improved sensitivity of nucleic acid database searches using application-specific scoring matrices. Methods Enzymol. 1991;3:66–70. [Google Scholar]
- Szymanski M., Barciszewska M.Z., Erdmann V.A., Barciszewski J., Barciszewska M.Z., Erdmann V.A., Barciszewski J., Erdmann V.A., Barciszewski J., Barciszewski J. 5S Ribosomal RNA Database. Nucleic Acids Res. 2002;30:176–178. doi: 10.1093/nar/30.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S., Hofacker I.L., Lukasser M., Hüttenhofer A., Stadler P.F., Hofacker I.L., Lukasser M., Hüttenhofer A., Stadler P.F., Lukasser M., Hüttenhofer A., Stadler P.F., Hüttenhofer A., Stadler P.F., Stadler P.F. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- Weinberg Z., Ruzzo W.L., Ruzzo W.L. Sequence-based heuristics for faster annotation of non-coding RNA families. Bioinformatics. 2006;22:445–452. doi: 10.1093/bioinformatics/bti743. [DOI] [PubMed] [Google Scholar]
- Workman C., Krogh A., Krogh A. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 1999;27:4816–4822. doi: 10.1093/nar/27.24.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J., de Van Peer Y., Winkelmans T., De Wachter R., de Van Peer Y., Winkelmans T., De Wachter R., Winkelmans T., De Wachter R., De Wachter R. The European database on small subunit ribosomal RNA. Nucleic Acids Res. 2002;30:183–185. doi: 10.1093/nar/30.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C. The uRNA database. Nucleic Acids Res. 1997;25:102–103. doi: 10.1093/nar/25.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]