Abstract
Autonomous non-long-terminal-repeat retrotransposons (NLRs) proliferate by retrotransposition via coordinated reactions of target DNA cleavage and reverse transcription by a mechanism called target-primed reverse transcription (TPRT). Whereas this mechanism guarantees the covalent attachment of the NLR and its target site at the 3′ junction, mechanisms for the joining at the 5′ junction have been conjectural. To better understand the retrotransposition pathways, we analyzed target–NLR junctions of zebrafish NLRs with a new method of identifying genomic copies that reside within other transposons, termed “target analysis of nested transposons” (TANT). Application of the TANT method revealed various features of the zebrafish NLR integrants; for example, half of the integrants carry extra nucleotides at the 5′ junction, which is in stark contrast to the major human NLR, LINE-1. Interestingly, in a cell culture assay, retrotransposition of the zebrafish NLR in heterologous human cells did not bear extra 5′ nucleotides, indicating that the choice of the 5′ joining pathway is affected by the host. Our results suggest that several pathways exist for NLR retrotransposition and argue in favor of host protein involvement. With genomic sequence information accumulating exponentially, our data demonstrate the general applicability of the TANT method for the analysis of a wide variety of retrotransposons.
Non-long-terminal-repeat retrotransposons (NLRs), including long interspersed nuclear elements (LINEs), comprise a substantial portion of many eukaryotic genomes (Arkhipova and Meselson 2000; Kazazian Jr. 2004). NLRs are divided into at least 11 clades, and the establishment of each clade dates back to the Precambrian era (Malik et al. 1999). Despite such ancient divergence, these elements share similar sequence features and mobility pathways. A typical intact NLR consists of a 5′-untranslated region (UTR), two open reading frames (ORFs), and a 3′ UTR with a microsatellite tail (Fig. 1). Most genomic NLR copies are truncated to various lengths in their 5′ regions. The product of the first ORF, ORF1p, typically exhibits RNA binding and nucleic acid chaperon activities and has been proposed to play a role in the stabilization of NLR RNA (Hohjoh and Singer 1996; Kolosha and Martin 1997; Martin and Bushman 2001; Martin et al. 2005a). The ORF2 protein, ORF2p, has two distinct enzymatic activities—endodeoxyribonuclease (EN) and reverse transcriptase (RT). During retrotransposition, ORF2p nicks the target duplex DNA and subsequently initiates reverse transcription of the NLR RNA using the 3′-OH end of the nicked DNA as a primer to synthesize an antisense-strand DNA of a new NLR copy by the mechanism called target-primed reverse transcription (TPRT) (Luan et al. 1993; Cost et al. 2002) (see Fig. 6, below). The second strand of the target duplex becomes cleaved during or after TPRT, detaching the upstream region of the target duplex from the downstream DNA. The sense-strand synthesis and joining of the NLR and upstream target DNA at the 5′ junction complete retrotransposition; however, detailed mechanisms for these steps remain speculative.
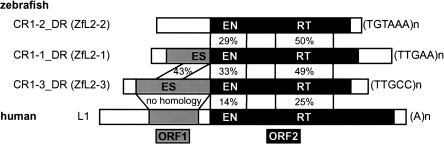
Figure 1.
Schematic representation of the NLRs analyzed in this study. ORF1 and ORF2 are shown in gray and black, respectively. The terminal repeats are shown on the right. The endonuclease, reverse transcriptase, and esterase domains are shown as EN, RT, and ES, respectively. The percentages indicate amino acid identities between any two proteins.
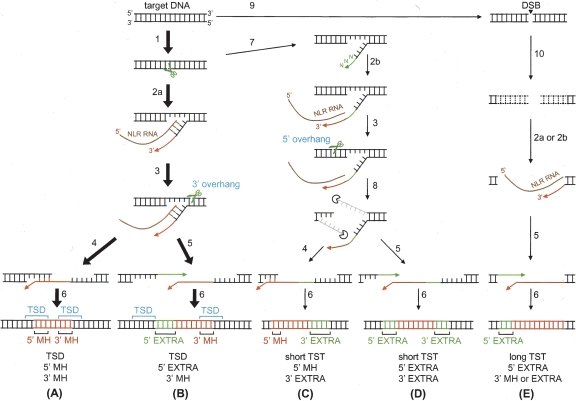
Figure 6.
Possible pathways for NLR retrotransposition. Major pathways that result in TSD and 3′ MH stretches are indicated by bold arrows (A,B), and the others are indicated by thin arrows (C,D,E). Some other pathways, although not shown, are possible; for example, one that generates long TSTs and 5′ MH. The numbered arrows indicate the following reactions: (1) first-strand cleavage by NLR-encoded ENs, (2a) reverse transcription initiated with the help of annealing of target DNA and NLR RNA, (2b) reverse transcription primed by extra nucleotides at the 3′ end, (3) second-strand cleavage, (4) annealing of nascent NLR cDNA and target DNA, (5) addition of extra 5′ nucleotides, (6) sense-strand synthesis and ligation, (7) addition of extra 3′ nucleotides, (8) nucleolytic digestion of overhanging sequences, (9) introduction of a double-strand break (DSB), and (10) exonucleolytic digestion from the DNA ends. (EXTRA) Extra nucleotides added either the 5′ or 3′ end.
Upon retrotransposition of the major mammalian NLRs, L1s, the target-site sequence of 8–20 base pairs (bp) is duplicated at each L1 end (i.e., target-site duplication, TSD) (Moran et al. 1996; Gilbert et al. 2002, 2005; Symer et al. 2002; Babushok et al. 2006). Thus, TSD enables us to determine the target-site boundaries of genomic L1 copies (Szak et al. 2002; Martin et al. 2005b; Zingler et al. 2005). At the 5′ junction, 5′-truncated L1 copies often share microhomology (MH) stretch with the end of the TSD, whereas full-length copies do not. These studies have led to models where the 5′ junction is joined via annealing of the NLR and target DNAs in the MH stretch (Feng et al. 1998; Martin et al. 2005b; Zingler et al. 2005). It also has been proposed that RTs jump from the RNA template to the target DNA via the MH stretch (George et al. 1996; Babushok et al. 2006).
In contrast to L1s, some NLRs are not associated with such obvious TSDs. Thus, reliable identification of the NLR-target junctions of their genomic copies remains difficult, although genomic sequence information has been accumulating in recent years. We considered that such difficulty could be overcome by collecting genomic NLR copies that reside within other transposons, because the preintegration sequence could be inferred from the consensus sequence of the host transposon. Hereafter, we refer to this collection strategy as target analysis of nested transposons, or TANT method.
The L2 clade of NLRs is represented by currently extinct LINE-2 (L2) in mammals, where the dominant active NLR clade is L1. The zebrafish genome harbors at least three active NLRs of the L2 clade: CR1–1_DR, CR1–2_DR, and CR1–3_DR, which are also called ZfL2–1, ZfL2–2, and ZfL2–3, respectively (Kapitonov and Jurka 2003; Sugano et al. 2006). CR1–1_DR and CR1–3_DR carry two ORFs. Their ORF2s encode an EN/RT protein and ORF1s encode an esterase-like protein (Fig. 1). Roles for ORF1p in retrotransposition are unknown, although mutations in ORF1 of CR1–1_DR decrease the frequency of retrotransposition (Sugano et al. 2006). CR1–2_DR carries only a single ORF, which encodes an EN/RT protein (Fig. 1). These three NLRs end with tandem repeats and apparently are not associated with a TSD (Kapitonov and Jurka 2003).
In this report, we used the TANT method to characterize genomic copies of these L2-clade NLRs. Bioinformatic analyses of their target-NLR junctions, in combination with the analysis of CR1–2_DR, experimentally retrotransposed in HeLa cells, revealed previously unrecognized consequences of their retrotransposition, and suggests the involvement of host functions in joining the NLR and target DNAs. Our data thus demonstrate the general applicability of the TANT method to the study of the mobility pathways of a wide variety of transposons.
Results
Collection of human L1s within transposons
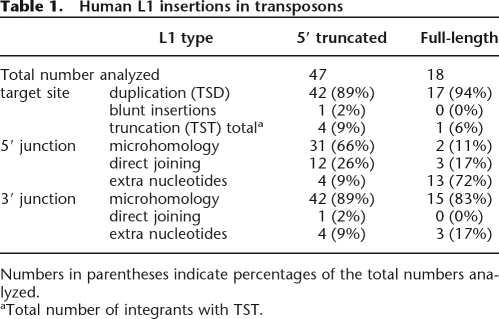
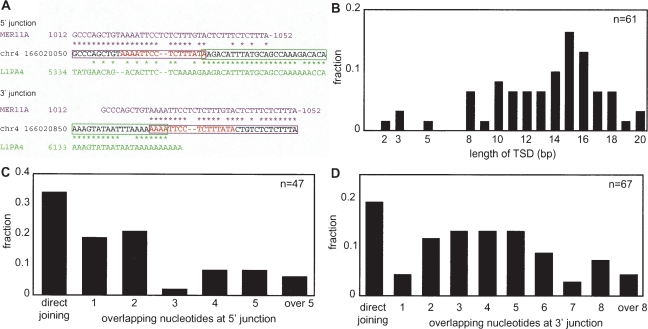
To test the effectiveness of the TANT method, we first collected genomic copies of human L1 that reside within other transposons, because target-site junctions of this NLR have been well characterized. For 47 5′-truncated and 18 full-length genomic copies analyzed, L1 copies are predominantly associated with TSDs of 8–20 bp and share a MH stretch with their target sites at the 3′ junctions (Table 1; Fig. 2A). At the 5′ junction, many of 5′-truncated copies (66%) have MH stretches and only a few copies (9%) are associated with an insertion of nucleotides of unknown origin, whereas many full-length L1s (72%) contain 2–16 extra nucleotides (Table 1). The length distributions of TSDs and MH stretches (Fig. 2B,C,D), target preference to 5′-TTAAAA-3′ (data not shown), and the discrete 5′ differences between 5′-truncated and full-length L1s are well consistent with previous observations of L1 copies collected and chosen based on the presence of an obvious TSD (Szak et al. 2002; Martin et al. 2005b; Zingler et al. 2005). Thus, the TANT method data reliably represent a general feature of target-NLR junctions generated by retrotransposition.
Table 1.
Human L1 insertions in transposons
Numbers in parentheses indicate percentages of the total numbers analyzed.
aTotal number of integrants with TST.
Figure 2.
Analysis of L1 integrants within other transposons. (A) An example of a genomic L1 integrant. Only junction regions are shown. The consensus sequence of the host transposon, MER11A (magenta), the sequence of a part of human chromosome 4 (black), and the L1PA4 consensus sequence (green) are aligned. Asterisks indicate identical sequences and the magenta and green boxes indicate MER11A and L1PA4 regions, respectively, inferred from the alignments. The target-site duplication (TSD) is shown in red letters. (B) Length distribution of TSD. Both full-length and 5′-truncated copies are included. (C) Length distribution of the 5′ microhomology (MH). Both full-length and 5′-truncated copies are included. (D) Length distribution of the 3′ MH. Both full-length and 5′-truncated copies are included.
Target sites of CR1_DRs within transposons
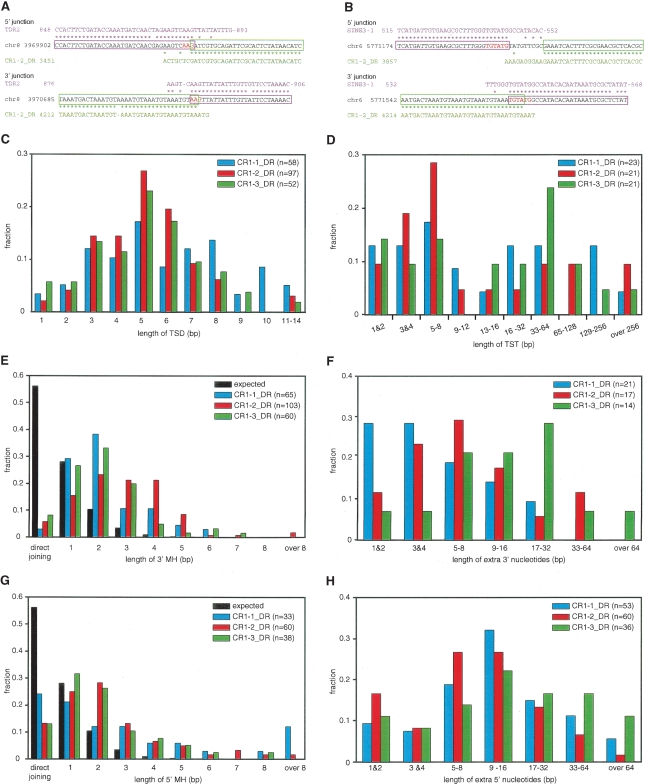
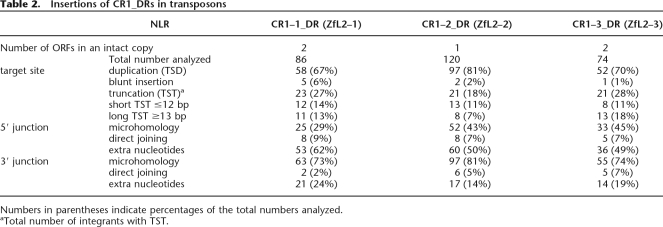
We next applied the TANT method to analyze CR1_DRs in the zebrafish genome sequence, because they are vertebrate non-L1 NLRs whose retrotransposition has been studied experimentally (Sugano et al. 2006). We collected 86, 120, and 74 copies of transposon-harbored CR1–1_DR, CR1–2_DR, and CR1–3_DR, respectively, with only a few being full-length insertions and the rest containing 5′ truncations. Analysis of their 5′ and 3′ junctions revealed that, in contrast to a previous report (Kapitonov and Jurka 2003), all three NLRs are predominantly (67%–81% of total) associated with TSDs of 1–14 bp (Fig. 3A,B for examples; Table 2 for statistics). The TSD lengths of all the three NLRs seem to follow a Gaussian distribution, with a mode of 5 bp and the majority between 3 and 8 bp (Fig. 3C).
Figure 3.
Analysis of integrants of CR1_DRs. (A,B) Two examples of CR1–2_DR integrants. The consensus sequences of the host transposons (magenta, TDR2 in A and SINE3–1 in B), the sequences of parts of the zebrafish genome (black), and the CR1–2_DR consensus sequence (green) are aligned. Asterisks indicate identical sequences, and the magenta and green boxes indicate the regions of CR1–2_DR and host transposons inferred from the alignments. The TSD is shown in red letters. The two integrants have TSDs and 3′ MHs. The example in A has 5′ MH, whereas the example in B has an insertion of extra 5′ nucleotides. (C,D) Length distributions of TSD (C) and TST (D) of CR1_DRs. The colors that discriminate each CR1_DR are indicated in the key in each panel, with the total numbers of CR1_DRs analyzed in parentheses. (E,G) Length distributions of MH at the 3′ (E) and 5′ (G) junctions. The probability distribution of the MH length for two random sequences was calculated as described previously (Roth et al. 1985) and is shown in black. The P values for expected and observed numbers were ≪0.01 in all χ2 tests. (F,H) Length distributions of extra nucleotides at the 3′ (F) and 5′ (H) junctions.
Table 2.
Insertions of CR1_DRs in transposons
Numbers in parentheses indicate percentages of the total numbers analyzed.
aTotal number of integrants with TST.
A very small fraction (1%–6%) is blunt inserted, whereas a larger fraction (18%–28%) is associated with target-site truncations (TSTs) (Table 2; Supplemental Fig. S1). The truncations range from 1 to 549 bp, as estimated by using the consensus sequences of the host transposons as a guide (Fig. 3D). Interestingly, they show a bimodal distribution, discriminating short and long TSTs (≤12 bp and ≥13 bp, respectively), suggesting that two different mechanisms underlie the target truncation upon retrotransposition (see Discussion).
For all CR1_DRs, compilation of target sequences around the insertion sites do not indicate any strong nucleotide preference at any position, although a downstream sequence of several base pairs is somewhat AT-rich in targets of CR1–1_DR and CR1–2_DR (Supplemental Fig. S2). This very weak preference could be explained by either some degree of cleavage specificity of NLR-encoded ENs or selection for cleavage products that can anneal with the NLR RNAs to start TPRT (see below). In any event, any nucleotide is allowed at almost all positions, leading to our conclusion that all CR1_DRs have very little sequence specificity for their integration targets.
Features of the 3′ junctions of CR1_DRs
Homology between the ends of the downstream target DNA and the NLR RNA to be reverse transcribed has been documented for L1s, suggesting that these homologous regions anneal to promote the TPRT initiation (Ostertag and Kazazian Jr. 2001; Kulpa and Moran 2006). Interestingly, 73%–81% of the integrants of CR1_DRs also show such MH between targets and the terminal tandem repeats of the integrating NLRs (Fig. 3A,B; Table 2; Supplemental Fig. S1). The lengths of the MH stretches differ significantly from that of two random sequences (Fig. 3E). L1- and CR1/L2-clades of NLRs diverged more than 400 million years ago (Malik et al. 1999), and they show different degrees of target-site specificity. Therefore, the conservation of the 3′ feature with MH suggests that annealing of target DNA and NLR RNA is a general mechanism to assist the TPRT initiation of many NLRs. In summary, the majority of the integrants (58%–71% of total) of CR1_DRs are associated with both a short TSD and 3′ MH (Supplemental Table S1).
On the other hand, we also found that some copies (14%–24%) are associated with an insertion of nucleotides (1–91 bp) at the 3′ junction (Fig. 3F; Table 2; Supplemental Fig. S1). Because we could not find the putative original NLR copies carrying the extra 3′ nucleotides in the current database, these extra 3′ nucleotides do not seem to be products of 3′ transduction, a retrotransposition event where the new copy is accompanied by the 3′-flanking region of the original copy. Thus, the extra 3′ nucleotides were presumably added after transcription of the NLR RNA. For one integrant, we found a genomic region (on a different chromosome) that is 87% identical to its extra 3′ nucleotides (61 bp). For seven examples of extra 3′ nucleotides (15–22 bp), we found homologous EST sequences (with 93%–100% identity). These extra nucleotides may have been generated by use of DNA or RNA templates. However, we could not find potential templates for seven of the 15 examples of the extra 3′ nucleotides that are 15 bp or longer. It is therefore likely that, in general, the extra 3′ nucleotides were generated by nontemplated DNA synthesis or by use of very short template regions. It has been reported that R2- and L1-encoded RTs can add nucleotides without a template or with a very short region(s) of a template before initiating the canonical RNA-templated TPRT (Luan and Eickbush 1995; Cost et al. 2002). The RTs encoded by CR1_DRs may also have such an activity that consequently generates extra 3′ nucleotides.
Features of the 5′ junctions of CR1_DRs
The 5′ junctions of the zebrafish NLRs show quite different features from those of L1. Whereas only a minor fraction of the genomic L1 integrants (9% of the total 5′-truncated copies) (Table 1) has extra nucleotides of unknown origin at the 5′ junction, about half (49%–62%) of the 5′-truncated copies of CR1_DRs contain such extra 5′ nucleotides. The two full-length insertions also contain extra 5′ nucleotides. The additions are 1–114 bp in length with an AT content of 65%–71% (Fig. 3B,H; Table 2; Supplemental Fig. S1). Because almost all of these elements are 5′ truncated and because we could not find their putative original NLR copies in the current database, it is unlikely that these integrants are products of 5′ transduction. Rather, the extra 5′ nucleotides were probably added after transcription of the NLR RNA. Thus, we searched for sequences in the database that are homologous to the respective sequences of the extra 5′nucleotides: we found five examples of genomic regions (42–99 bp with 85%–92% identity) and three examples of EST sequences (15–50 bp with 90%–100% identity). However, we could not find any such homologous sequences in the current database for most (47 of 54) of the extra nucleotides that are ≥15 bp. These suggest that the extra 5′ nucleotides were, in general, created in a nontemplated manner or via switching very short template regions (i.e., template switching).
Some of the integrants (29%–45%) had MH at the 5′ junctions (Fig. 3A; Table 2 Supplemental Fig. S1). The length distributions of these MHs differ significantly from that expected for two random sequences (Fig. 3G). Thus, the MHs may not have been generated by chance, but by a mechanism integral to certain retrotransposition pathways (see Discussion). In summary, CR1_DRs carry either extra nucleotides or MH at their 5′ junctions.
Insertion of extra 3′ nucleotides and truncation of target sites are inter-related
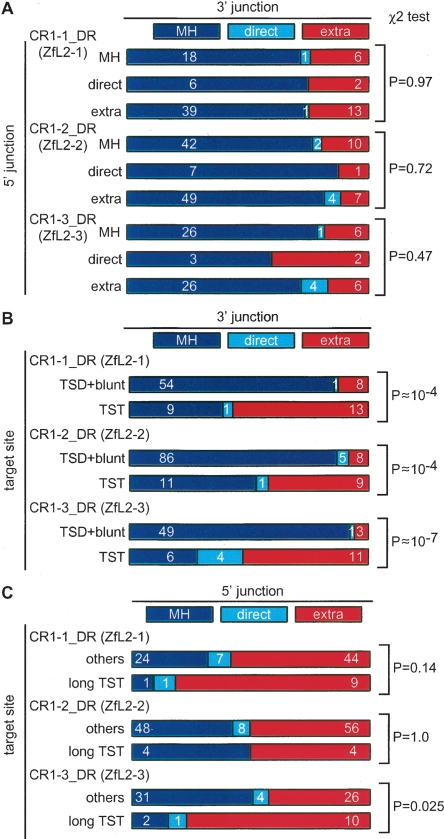
We analyzed the relationships among the features of target-site alterations and 5′ and 3′ junctions. We did not find a significant relationship between the features of 3′ and 5′ junctions (Fig. 4A). However, statistical analyses revealed that target-site alterations and the features of 3′ junctions are interrelated (P |LL 0.01) (Fig. 4B). Indeed, insertion of the extra 3′ nucleotides is enriched in integrants with TSTs (Fig. 4B): about half (48%–57%) of TST integrants are associated with insertion of the extra 3′ nucleotides, suggesting their mechanistic inter-relationship (see Discussion).
Figure 4.
Two-dimensional matrix analysis for the interrelatedness of the junction features. (A) 5′ junction vs. 3′ junction. Integrants are categorized into those having MH, direct joining (direct), or extra nucleotides (extra) at the 5′ junction. In each category, integrants are further categorized into those having MH (blue), direct joining (light blue), and extra nucleotides (red) at the 3′ junction. The number of copies collected is indicated inside each rectangle and the P values by χ2 tests of independence are shown at the right. (B) Target-site alterations vs. 3′ junction. Integrants are categorized into those with TSTs and the others (TSD+blunt), and further categorized by their 3′ features. The number of copies collected is indicated inside each rectangle. (C) Target-site alterations vs. 5′ junction. Integrants are categorized into those with long TSTs or others (short TSTs, TSDs, or blunt insertion), and further categorized by their 5′ features. The number of copies collected is indicated inside. Integrants with MH and those joined directly were combined for the χ2 analysis to enhance the power of validation.
Copies of the dual-ORF CR1_DRs with long TSTs are biased toward insertion of extra 5′ nucleotides
Finally, we analyzed the relationship between the 5′ junction and target-site alterations (Fig. 4C; Supplemental Table S2). We found some interrelation: Most long TST-associated integrants of CR1–1_DR and CR1–3_DR (82% and 77%, respectively) carry the extra 5′ nucleotides. On the other hand, we did not find any interrelation for the CR1–2_DR integrants. It is worth considering that CR1–2_DR has a single ORF, whereas CR1–1_DR and CR1–3_DR have two ORFs (Fig. 1). Thus, although the exact mechanism(s) for generation of the extra 5′ nucleotides is unknown at present, the ORF1 proteins may be involved in the pathways for joining the 5′ junction during a type of retrotransposition where target sites suffer extensive truncation (see Discussion).
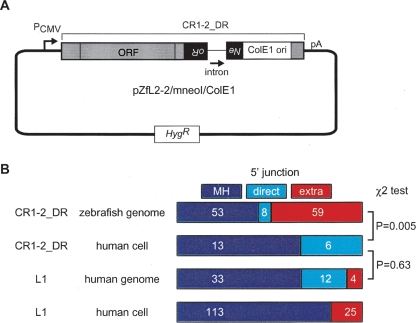
Retrotransposition of CR1–2_DR in HeLa cells does not create integrants with extra 5′ nucleotides
As described above, zebrafish CR1_DRs and human L1 have significantly different outcomes with regard to the features of the 5′ junction. Because different NLRs retrotransposed in different hosts were compared, it remained unknown whether this discrepancy could be ascribed to differences in the NLR-encoded proteins or in the hosts. To address this question, we had a genetically marked CR1–2_DR (Fig. 5A) retrotranspose in cultured human cells. Of the 19 integrants we determined, none carried extra nucleotides at the 5′ junction, whereas 13 clones (72%) had MH and others were directly jointed (Fig. 5B). This is in striking contrast with CR1–2_DR integrations in the zebrafish genome, and instead resembles L1 insertions in human (Fig. 5B). Therefore, it seems likely that the patterns of the 5′ joining are dictated by the host.
Figure 5.
Retrotransposition assay in HeLa cells. (A) Construction of pZfL2–2/mneoI/ColE1. A full-length CR1–2_DR was placed under the control of the CMV promoter (PCMV) in the pCEP4 vector carrying the hygromycin-resistance gene (Hyg). The mneoI marker and ColE1 origin were inserted in the 3′ UTR of CR1–2_DR. The mneoI marker is a neomycin-resistant gene (Neo) interrupted by an insertion of an intron in the antisense orientation. This marker itself is in an antisense orientation relative to the NLR transcript. Thus, the vector does not confer the Neo phenotype, whereas CR1–2_DR retrotransposition, which includes splicing of the transcript, reverse transcription of that spliced RNA, and insertion of the synthesized cDNA into the genomic DNA, restores an intact Neo gene, converting the host cell to Neo. (B) Statistics for various NLR integrants. Integrants of CR1–2_DR in the zebrafish genome (top), those in HeLa cells using pZfL2–2/mneoI/ColE1 (second), L1 copies in transposons in the human genome (third), and de novo L1 insertions in human cultured cells (bottom) were categorized with regard to 5′ junctions. The frequency of extra 5′ nucleotides in de novo L1 insertions were reported previously (Symer et al. 2002; Gilbert et al. 2005). Because MH and direct joining were not distinguished in these reports, these integrants are represented as MH. The number of copies collected is indicated inside each rectangle. The P-values by χ2 tests for each pair are indicated at the right.
Discussion
The TANT method for genome-wide analysis of transposon integrants
We developed the TANT method for large-scale analysis of boundaries of genomic NLR copies. This method takes advantage of the fact that we can mine genomic databases for NLR copies residing in other transposons and for consensus sequences of transposons; we can then use the information to determine the junction sites of nested NLRs. It is formally possible that secondary DNA rearrangements could have occurred at these junctions after retrotransposition, leading to misinterpretation. This possibility can, however, be minimized by selecting younger elements. Indeed, the statistics for the L1 elements collected and analyzed by the TANT method (Table 1) are very consistent with previous reports, thereby validating the method. Given that genomic sequence information for many kinds of higher eukaryotes has been expanding steadily, the TANT method will be generally applicable to investigation of many kinds of transposons, as we have shown here for L1 and CR1_DRs.
Retrotransposition of CR1_DRs predominantly generates a short TSD
Whereas it has been reported that CR1_DRs lack obvious TSDs (Kapitonov and Jurka 2003), our analysis reveals that the majority of these elements have TSDs (Table 2). The prevalence of TSD retention in the genomic copies we identified argues against the possibility of secondary rearrangements, as discussed above. Furthermore, the Gaussian-like distributions of the TSD lengths (Fig. 3C) imply that the first- and second-strand cleavages are ordered enzymatic reactions with the second-strand nick positioned downstream of the first-strand nick (Fig. 6A,B). Such ordered reactions have been proposed for R2 retrotransposition, in which the second strand is cleaved by a subunit of the (EN/RT)2 homodimer, whereas the other subunit is responsible for the first-strand cleavage and cDNA synthesis (Christensen and Eickbush 2005).
Target-site truncations as the outcome of noncanonical TPRT reactions
A fraction (18%–28%) of the integrants we identified is associated with TSTs (Table 2). Careful analysis revealed bimodal distributions of the TST lengths (Fig. 3D). For short TSTs, it has been proposed that nicking of the second strand at several base pairs upstream of the first nicking site and subsequent use of that nick to prime the sense-strand synthesis results in loss of the segment between the two nicks (Gilbert et al. 2002). If CR1_DRs use the ordered cleavages and reverse transcription discussed above, then how or when can such upstream second-strand cleavage compete over the canonical downstream nicks? A hint seems to lie in our observation that integrants with TSTs are biased toward the insertion of extra 3′ nucleotides (Fig. 4B). This coincidence can be explained by the assumption that a TPRT that starts with NLR RNA-independent DNA synthesis uncouples TPRT from the second-strand cleavage, thereby resulting in cleavage at unusual sites (Fig. 6C,D).
Implications for NLR-mediated DNA end joining
The upstream nicking model for creation of TSTs requires strand separation between the two nicked sites. However, the long TSTs include truncations of up to 0.5 kb; therefore, it is unlikely that they are products of this pathway. Rather, it is more likely that exonucleolytic digestion of the target duplex from the site of cleavage is involved in creating long TSTs. We note that loss of a relatively long DNA region resembles the DNA truncation seen after repair of double-strand breaks (DSBs) via nonhomologous DNA end joining (NHEJ), the major pathway for DSB repair in vertebrates (Lieber et al. 2003). Thus, it is tempting to speculate that the sequences of CR1_DRs can be captured at sites of DSB. In this model, the DNA repair machinery digests the target DNA, to some extent, from the DSB site. Although an RT-independent, DNA-based mechanism cannot be ruled out, we propose that the NLR RTs can use the sites of DSB to prime the reverse transcription for some fraction of events (Fig. 6E). This possibility has been proposed for mammalian L1s as well (Edgell et al. 1987; Morrish et al. 2002). It also has been reported that sequences of an LTR-retrotransposons (IAP) and a SINE (B1) were captured at DSB sites (Lin and Waldman 2001). These observations suggest that use of retrotransposon sequences to heal DSBs is a general paradigm.
Pathways for joining the 5′ junction are dictated by the host environments
The majority of L1 retrotransposition bear an MH stretch at the 5′ junction (Gilbert et al. 2002, 2005; Symer et al. 2002; Martin et al. 2005b; Zingler et al. 2005; Babushok et al. 2006). Here we showed the presence of such 5′ MH stretches in a substantial proportion of CR1_DRs integrants (Table 2). During these retrotranspositions, the target DNA (in most cases, the 3′ overhang generated by the second-strand nick) and the nascent NLR cDNA may have become annealed, and the 3′ end of the target strand may have been used to prime sense-strand synthesis (Fig. 6A,C), as proposed for L1 and R1 retrotranspositions, with the synthesis being catalyzed by NLR RTs (Feng et al. 1998; Martin et al. 2005b) or by a host DNA polymerase(s) (Zingler et al. 2005).
The most surprising finding in this study is that half or more of the CR1_DRs integrants carry extra 5′ nucleotides (Table 2). The nucleotides were likely added without a template during retrotransposition. The nucleotides could be synthesized from the 3′ end of nascent NLR cDNA generated by incomplete reverse transcription, as proposed by Babushok et al. (2006). Instead, we favor a model in which the nucleotides are synthesized from the 3′ end of the top strand of the target, while the RTs synthesize the long cDNA (Fig. 6B,D,E). We speculate that the extra 5′ nucleotides can in turn serve as a primer to synthesize the sense-strand DNA from somewhere on the nascent antisense-strand cDNA, producing integrants having 5′ truncations and extra 5′ nucleotides. In addition, CR1–1_DR and CR1–3_DR relatively frequently use this 5′ nucleotide-adding pathway in retrotransposition that results in a long TST (Fig. 4C). Because ORF1p of CR1–1_DR has DNA-binding activity (M. Nakamura, M. Kajikawa, and N. Okada, unpubl.), it may coat the nascent cDNA, which consequently prevents the target–cDNA annealing, thereby providing a better opportunity for addition of extra nucleotides.
Formally, the extra 5′ nucleotides could be synthesized by either NLR RTs or host DNA polymerases. If they were synthesized by NLR RTs, the tendency of the 5′-junction features would not be altered by changing the host. Unlike copies in the zebrafish genome, however, no CR1–2_DR integrant that experimentally retrotransposed in human cells carried extra 5′ nucleotides (Fig. 5). Rather, all integrants were associated with an MH stretch or direct joining, resembling the statistics of L1 retrotransposed in human. These results suggest that alternative pathways account for the integrants associated with MH at the 5′ junction and those with extra 5′ nucleotides, and that the pathway utilization is directed by the host. It may be possible that the endogenous L1 proteins affected CR1–2_DR retrotransposition pathways in HeLa cells, but is less likely because it does not explain why 5′ features are different between full-length and 5′-truncated L1 insertions in human. Rather, host factors are likely involved in the processes for joining of the target and NLR DNAs at the 5′ junction. Consistent with this idea, we have recently revealed that zebrafish L1-clade NLRs are approximately three times more frequently associated with extra 5′ nucleotides than human L1s are (Ichiyanagi and Okada 2006). Interestingly, it has been reported that several host-encoded DNA repair proteins are involved in the mobility of a bacterial group II intron (Smith et al. 2005), which retrotransposes and retrohomes via TPRT (Saldanha et al. 1999), and that the pathway utilization for its retrotransposition is dictated by the host (Coros et al. 2005). Recently, human L1 retrotransposition was suggested to require the activity of the ATM protein (Gasior et al. 2006), a central regulator of DNA-damage response (Shiloh and Kastan 2001), which is consistent with the idea that NLR retrotransposition intermediates are recognized as DNA damage and subsequently processed by DNA-repair proteins (Gilbert et al. 2005). We note that both MH and extra nucleotides are also seen in NHEJ-mediated repair products (Roth et al. 1989; Gottlich et al. 1998; Kabotyanski et al. 1998; Smith et al. 2003), hinting the involvement of the NHEJ machinery in 5′ joining during NLR retrotransposition. Further studies, including experiments using mutant hosts, will elucidate the exact pathways and factors involved in the amplification of these genomic symbionts.
Methods
Collection of transposon-harboring NLR copies and identification of target–NLR junctions
We downloaded RepeatMasker tables of interspersed repeats for human (hg17, May 2004) and zebrafish (danRer2, June 2004) genomes from the UCSC Genome Browser (Hinrichs et al. 2006). Using the tables and Perl scripts (available upon request), we screened L1 and CR1_DRs copies that reside within other host transposons using the following criteria: (1) NLR copies contain the complete 3′ region, (2) NLR copies sandwiched between two fragments of the same transposon with the same orientation (NLRs within a single transposon), (3) the divergences of the host-transposon sequences from the consensus sequences are <12%, (4) these values of divergence of the two host transposon fragments do not differ significantly (P > 0.05 by a χ2 test), (5) the insertions and deletions of the host transposons in comparison with the consensus sequences are <10%, (6) both fragments of the host transposon are >50 bp, and (7) the ends of the host transposon and NLR sequences are located <200 bp apart at both junctions. For 5′-truncated L1s, we selected integrants where L1 copies showed ≤3.8% divergence from the consensus sequences. For full-length L1s, we selected integrants where L1 copies showed ≤5.1% divergence. When duplicated fragments were collected, we used and counted only one of them. Complete data sets are available in the Supplemental files (L1.txt, CR1–1_DR.txt, CR1–2_DR.txt, and CR1–3_DR.txt).
The genomic sequence files were downloaded from the UCSC Browser, and consensus sequences of NLRs and host transposons were obtained from RepBase (Jurka et al. 2005). With the help of homology alignments, we manually analyzed the junctions of all NLR copies collected. To be conservative, we regarded MH as a segment with 100% identity between the target and NLR ends.
Construction of the CR1–2_DR vector, retrotransposition in HeLa cells, and sequence analysis
We amplified the region containing the mneoI retrotransposition marker and the ColE1 origin in pCEP4/L1.3mneoI/ColE1 (Gilbert et al. 2002) by PCR using a 5′ primer containing a NotI site and a 3′ primer containing a BamHI site. The PCR fragment was used to replace the NotI–BamHI fragment of pBB4 (Sugano et al. 2006), which carries the ORF of CD1–2_DR (clone ZL15) and a NotI–BamHI fragment containing the mneoI marker. The resulting plasmid was digested with BamHI and ligated with another PCR fragment that contains the 3′-tail region of ZL15 with BamHI sites at both ends. The resulting plasmid that carried the 3′ tail in the correct orientation was designated as pZfL2–2/mneoI/ColE1. Thus, this vector consisted of a CMV promoter, the CR1–2_DR ORF, mneoI, ColE1 ori, the 3′ tail of CR1–2_DR, and the polyadenylation signal in pCEP4 (Invitrogen) (Fig. 5). The mneoI marker allows selection (G418 resistance) of cultured cells that have undergone retrotransposition, and the ColE1 origin facilitates recovery of genomic fragments containing retrotransposed elements by self-circularization, transformation into Escherichia coli, and subsequent selection for self-replicating plasmids that confer kanamycin resistance. Isolation of cells carrying a retrotransposition product and determination of sequences of the integrants were performed as described previously (Gilbert et al. 2002) using primers specific to CR1–2_DR.
Acknowledgments
We thank Dr. John V. Moran for providing the vector, pCEP4/L1.3mneoI/ColE1, and for critical reading of the manuscript. We also thank Drs. Daria Babushok, Marlene Belfort, Jose L. Garcia-Perez, Haig H. Kazazian Jr., Mitsuhiro Nakamura, and Jun Suzuki for helpful comments on the manuscript. This work was supported by a Grant-in-Aid to N.O. from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the 21st Century Center of Excellence (COE) program of the ministry.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5542607
References
- Arkhipova I., Meselson M., Meselson M. Transposable elements in sexual and ancient asexual taxa. Proc. Natl. Acad. Sci. 2000;97:14473–14477. doi: 10.1073/pnas.97.26.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok D.V., Ostertag E.M., Courtney C.E., Choi J.M., Kazazian H.H., Jr., Ostertag E.M., Courtney C.E., Choi J.M., Kazazian H.H., Jr., Courtney C.E., Choi J.M., Kazazian H.H., Jr., Choi J.M., Kazazian H.H., Jr., Kazazian H.H., Jr. L1 integration in a transgenic mouse model. Genome Res. 2006;16:240–250. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.M., Eickbush T.H., Eickbush T.H. R2 target-primed reverse transcription: Ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol. Cell. Biol. 2005;25:6617–6628. doi: 10.1128/MCB.25.15.6617-6628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coros C.J., Landthaler M., Piazza C.L., Beauregard A., Esposito D., Perutka J., Lambowitz A.M., Belfort M., Landthaler M., Piazza C.L., Beauregard A., Esposito D., Perutka J., Lambowitz A.M., Belfort M., Piazza C.L., Beauregard A., Esposito D., Perutka J., Lambowitz A.M., Belfort M., Beauregard A., Esposito D., Perutka J., Lambowitz A.M., Belfort M., Esposito D., Perutka J., Lambowitz A.M., Belfort M., Perutka J., Lambowitz A.M., Belfort M., Lambowitz A.M., Belfort M., Belfort M. Retrotransposition strategies of the Lactococcus lactis Ll.LtrB group II intron are dictated by host identity and cellular environment. Mol. Microbiol. 2005;56:509–524. doi: 10.1111/j.1365-2958.2005.04554.x. [DOI] [PubMed] [Google Scholar]
- Cost G.J., Feng Q., Jacquier A., Boeke J.D., Feng Q., Jacquier A., Boeke J.D., Jacquier A., Boeke J.D., Boeke J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgell M.H., Hardies S.C., Loeb D.D., Shehee W.R., Padgett R.W., Burton F.H., Comer M.B., Casavant N.C., Funk F.D., Hutchison C.A., III, Hardies S.C., Loeb D.D., Shehee W.R., Padgett R.W., Burton F.H., Comer M.B., Casavant N.C., Funk F.D., Hutchison C.A., III, Loeb D.D., Shehee W.R., Padgett R.W., Burton F.H., Comer M.B., Casavant N.C., Funk F.D., Hutchison C.A., III, Shehee W.R., Padgett R.W., Burton F.H., Comer M.B., Casavant N.C., Funk F.D., Hutchison C.A., III, Padgett R.W., Burton F.H., Comer M.B., Casavant N.C., Funk F.D., Hutchison C.A., III, Burton F.H., Comer M.B., Casavant N.C., Funk F.D., Hutchison C.A., III, Comer M.B., Casavant N.C., Funk F.D., Hutchison C.A., III, Casavant N.C., Funk F.D., Hutchison C.A., III, Funk F.D., Hutchison C.A., III, Hutchison C.A., III The L1 family in mice. Prog. Clin. Biol. Res. 1987;251:107–129. [PubMed] [Google Scholar]
- Feng Q., Schumann G., Boeke J.D., Schumann G., Boeke J.D., Boeke J.D. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc. Natl. Acad. Sci. 1998;95:2083–2088. doi: 10.1073/pnas.95.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S.L., Wakeman T.P., Xu B., Deininger P.L., Wakeman T.P., Xu B., Deininger P.L., Xu B., Deininger P.L., Deininger P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006;375:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J.A., Burke W.D., Eickbush T.H., Burke W.D., Eickbush T.H., Eickbush T.H. Analysis of the 5′ junctions of R2 insertions with the 28S gene: Implications for non-LTR retrotransposition. Genetics. 1996;142:853–863. doi: 10.1093/genetics/142.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N., Lutz-Prigge S., Moran J.V., Lutz-Prigge S., Moran J.V., Moran J.V. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Gilbert N., Lutz S., Morrish T.A., Moran J.V., Lutz S., Morrish T.A., Moran J.V., Morrish T.A., Moran J.V., Moran J.V. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlich B., Reichenberger S., Feldmann E., Pfeiffer P., Reichenberger S., Feldmann E., Pfeiffer P., Feldmann E., Pfeiffer P., Pfeiffer P. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. Eur. J. Biochem. 1998;258:387–395. doi: 10.1046/j.1432-1327.1998.2580387.x. [DOI] [PubMed] [Google Scholar]
- Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Diekhans M., Furey T.S., Harte R.A., Hsu F., Furey T.S., Harte R.A., Hsu F., Harte R.A., Hsu F., Hsu F., et al. The UCSC Genome Browser Database: Update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H., Singer M.F., Singer M.F. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- Ichiyanagi K., Okada N., Okada N. Genomic alterations upon integration of zebrafish L1 elements revealed by the TANT method. Gene. 2006;383:108–116. doi: 10.1016/j.gene.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Klonowski P., Kohany O., Walichiewicz J., Kohany O., Walichiewicz J., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kabotyanski E.B., Gomelsky L., Han J.O., Stamato T.D., Roth D.B., Gomelsky L., Han J.O., Stamato T.D., Roth D.B., Han J.O., Stamato T.D., Roth D.B., Stamato T.D., Roth D.B., Roth D.B. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J., Jurka J. The esterase and PHD domains in CR1-like non-LTR retrotransposons. Mol. Biol. Evol. 2003;20:38–46. doi: 10.1093/molbev/msg011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H.H., Jr. Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kolosha V.O., Martin S.L., Martin S.L. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl. Acad. Sci. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa D.A., Moran J.V., Moran J.V. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- Lieber M.R., Ma Y., Pannicke U., Schwarz K., Ma Y., Pannicke U., Schwarz K., Pannicke U., Schwarz K., Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- Lin Y., Waldman A.S., Waldman A.S. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158:1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D.D., Eickbush T.H., Eickbush T.H. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H., Korman M.H., Jakubczak J.L., Eickbush T.H., Jakubczak J.L., Eickbush T.H., Eickbush T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Malik H.S., Burke W.D., Eickbush T.H., Burke W.D., Eickbush T.H., Eickbush T.H. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- Martin S.L., Bushman F.D., Bushman F.D. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.L., Cruceanu M., Branciforte D., Wai-Lun Li P., Kwok S.C., Hodges R.S., Williams M.C., Cruceanu M., Branciforte D., Wai-Lun Li P., Kwok S.C., Hodges R.S., Williams M.C., Branciforte D., Wai-Lun Li P., Kwok S.C., Hodges R.S., Williams M.C., Wai-Lun Li P., Kwok S.C., Hodges R.S., Williams M.C., Kwok S.C., Hodges R.S., Williams M.C., Hodges R.S., Williams M.C., Williams M.C. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J. Mol. Biol. 2005a;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Martin S.L., Li W.L., Furano A.V., Boissinot S., Li W.L., Furano A.V., Boissinot S., Furano A.V., Boissinot S., Boissinot S. The structures of mouse and human L1 elements reflect their insertion mechanism. Cytogenet. Genome Res. 2005b;110:223–228. doi: 10.1159/000084956. [DOI] [PubMed] [Google Scholar]
- Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr., Boeke J.D., Kazazian H.H., Jr., Kazazian H.H., Jr. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Morrish T.A., Gilbert N., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V., Gilbert N., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V., Taccioli G.E., Batzer M.A., Moran J.V., Batzer M.A., Moran J.V., Moran J.V. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- Ostertag E.M., Kazazian H.H., Jr., Kazazian H.H., Jr. Twin priming: A proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D.B., Porter T.N., Wilson J.H., Porter T.N., Wilson J.H., Wilson J.H. Mechanisms of nonhomologous recombination in mammalian cells. Mol. Cell. Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D.B., Chang X.B., Wilson J.H., Chang X.B., Wilson J.H., Wilson J.H. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol. Cell. Biol. 1989;9:3049–3057. doi: 10.1128/mcb.9.7.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha R., Chen B., Wank H., Matsuura M., Edwards J., Lambowitz A.M., Chen B., Wank H., Matsuura M., Edwards J., Lambowitz A.M., Wank H., Matsuura M., Edwards J., Lambowitz A.M., Matsuura M., Edwards J., Lambowitz A.M., Edwards J., Lambowitz A.M., Lambowitz A.M. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry. 1999;38:9069–9083. doi: 10.1021/bi982799l. [DOI] [PubMed] [Google Scholar]
- Shiloh Y., Kastan M.B., Kastan M.B. ATM: Genome stability, neuronal development, and cancer cross paths. Adv. Cancer Res. 2001;83:209–254. doi: 10.1016/s0065-230x(01)83007-4. [DOI] [PubMed] [Google Scholar]
- Smith J., Riballo E., Kysela B., Baldeyron C., Manolis K., Masson C., Lieber M.R., Papadopoulo D., Jeggo P., Riballo E., Kysela B., Baldeyron C., Manolis K., Masson C., Lieber M.R., Papadopoulo D., Jeggo P., Kysela B., Baldeyron C., Manolis K., Masson C., Lieber M.R., Papadopoulo D., Jeggo P., Baldeyron C., Manolis K., Masson C., Lieber M.R., Papadopoulo D., Jeggo P., Manolis K., Masson C., Lieber M.R., Papadopoulo D., Jeggo P., Masson C., Lieber M.R., Papadopoulo D., Jeggo P., Lieber M.R., Papadopoulo D., Jeggo P., Papadopoulo D., Jeggo P., Jeggo P. Impact of DNA ligase IV on the fidelity of end joining in human cells. Nucleic Acids Res. 2003;31:2157–2167. doi: 10.1093/nar/gkg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D., Zhong J., Matsuura M., Lambowitz A.M., Belfort M., Zhong J., Matsuura M., Lambowitz A.M., Belfort M., Matsuura M., Lambowitz A.M., Belfort M., Lambowitz A.M., Belfort M., Belfort M. Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming. Genes & Dev. 2005;19:2477–2487. doi: 10.1101/gad.1345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano T., Kajikawa M., Okada N., Kajikawa M., Okada N., Okada N. Isolation and characterization of retrotransposition-competent LINEs from zebrafish. Gene. 2006;365:74–82. doi: 10.1016/j.gene.2005.09.037. [DOI] [PubMed] [Google Scholar]
- Symer D.E., Connelly C., Szak S.T., Caputo E.M., Cost G.J., Parmigiani G., Boeke J.D., Connelly C., Szak S.T., Caputo E.M., Cost G.J., Parmigiani G., Boeke J.D., Szak S.T., Caputo E.M., Cost G.J., Parmigiani G., Boeke J.D., Caputo E.M., Cost G.J., Parmigiani G., Boeke J.D., Cost G.J., Parmigiani G., Boeke J.D., Parmigiani G., Boeke J.D., Boeke J.D. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- Szak S.T., Pickeral O.K., Makalowski W., Boguski M.S., Landsman D., Boeke J.D., Pickeral O.K., Makalowski W., Boguski M.S., Landsman D., Boeke J.D., Makalowski W., Boguski M.S., Landsman D., Boeke J.D., Boguski M.S., Landsman D., Boeke J.D., Landsman D., Boeke J.D., Boeke J.D. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-research0052. research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingler N., Willhoeft U., Brose H.P., Schoder V., Jahns T., Hanschmann K.M., Morrish T.A., Lower J., Schumann G.G., Willhoeft U., Brose H.P., Schoder V., Jahns T., Hanschmann K.M., Morrish T.A., Lower J., Schumann G.G., Brose H.P., Schoder V., Jahns T., Hanschmann K.M., Morrish T.A., Lower J., Schumann G.G., Schoder V., Jahns T., Hanschmann K.M., Morrish T.A., Lower J., Schumann G.G., Jahns T., Hanschmann K.M., Morrish T.A., Lower J., Schumann G.G., Hanschmann K.M., Morrish T.A., Lower J., Schumann G.G., Morrish T.A., Lower J., Schumann G.G., Lower J., Schumann G.G., Schumann G.G. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res. 2005;15:780–789. doi: 10.1101/gr.3421505. [DOI] [PMC free article] [PubMed] [Google Scholar]