Abstract
Chlamydia trachomatis is an obligate intracellular bacterium of major public health significance, infecting over one-tenth of the world’s population and causing blindness and infertility in millions. Mounting evidence supports recombination as a key source of genetic diversity among free-living bacteria. Previous research shows that intracellular bacteria such as Chlamydiaceae may also undergo recombination but whether this plays a significant evolutionary role has not been determined. Here, we examine multiple loci dispersed throughout the chromosome to determine the extent and significance of recombination among 19 laboratory reference strains and 10 present-day ocular and urogenital clinical isolates using phylogenetic reconstructions, compatibility matrices, and statistically based recombination programs. Recombination is widespread; all clinical isolates are recombinant at multiple loci with no two belonging to the same clonal lineage. Several reference strains show nonconcordant phylogenies across loci; one strain is unambiguously identified as recombinantly derived from other reference strain lineages. Frequent recombination contrasts with a low level of point substitution; novel substitutions relative to reference strains occur less than one per kilobase. Hotspots for recombination are identified downstream from ompA, which encodes the major outer membrane protein. This widespread recombination, unexpected for an intracellular bacterium, explains why strain-typing using one or two genes, such as ompA, does not correlate with clinical phenotypes. Our results do not point to specific events that are responsible for different pathogenicities but, instead, suggest a new approach to dissect the genetic basis for clinical strain pathology with implications for evolution, host cell adaptation, and emergence of new chlamydial diseases.
Chlamydia trachomatis is the primary bacterial cause of preventable blindness and sexually transmitted diseases (STD) worldwide (World Health Organization 2001; Dean 2002). The costs for productivity loss in underdeveloped countries and treatment in the developed world amount to billions of dollars annually (Frick et al. 2003; Centers for Disease Control and Prevention 2005). Over 600 million people worldwide are infected, with seven million blind and 100 million suffering severe visual deficits (Dean 2002). About four million new urogenital infections occur annually in the United States (Centers for Disease Control and Prevention 2005). Many are asymptomatic and can recur, constituting a large reservoir of untreated individuals who can transmit the organism and develop sequelae such as infertility and life-threatening ectopic pregnancy (Centers for Disease Control and Prevention 2005). C. trachomatis also increases the risk for invasive squamous-cell carcinoma of the cervix (Anttila et al. 2001) and HIV-1 transmission (Lavreys et al. 2004).
C. trachomatis serotyping is based on the antigenic major outer membrane protein (MOMP), and differentiates 19 serovars or strains (including genovariant Ja), which are grouped into three seroclasses: B Class (B, Ba, D, Da, E, L1, L2, L2a); C Class (A, C, H, I, Ia, J, Ja, K, L3), and Intermediate Class (F and G) (Wang and Grayston 1991a, b). Sequencing of the ompA gene that encodes MOMP has distinguished subtypes of these reference strains (Dean et al. 1992; Brunham et al. 1994; Hayes et al. 1994; Dean and Millman 1997). Yet, no typing scheme differentiates the three C. trachomatis disease groups: ocular (trachoma; reference strains A, B, Ba, C), urogenital (reference strains D–K, Da, Ia, Ja), and invasive urogenital (lymphogranuloma venereum [LGV]; reference strains L1, L2, L2a, L3). For example, not all strains that cause chronic ocular trachoma belong to the same seroclass (Dean 2002); this is also the case for the two other disease groups (Boisvert et al. 1999; Millman et al. 2004).
Relatively little is known about the C. trachomatis genetic factors that may be involved in tissue tropism, pathogenesis, and evolution. This is largely due to its intracellular nature and the inability to genetically manipulate the organism. However, genome sequencing of two of the 19 reference strains of C. trachomatis, D/UW-3 (Stephens et al. 1998) and A/Har-13 (Carlson et al. 2005), has provided some knowledge of genes such as the cytotoxin locus (Carlson et al. 2004), trpR, trpB, trpA, and trpC of the partial tryptophan biosynthesis operon (Caldwell et al. 2003) and the nine-member polymorphic membrane protein (Pmp) gene (pmp) family (Stothard et al. 2003; Gomes et al. 2006), the latter of which are unique to the family Chlamydiaceae, that may be important in tissue tropism or pathogenesis. Phylogenetic studies of partial pmp sequences of 15 C. trachomatis reference strains (Stothard et al. 2003) and complete pmp sequences of the 19 reference strains (Gomes et al. 2006) demonstrated genetic clustering by disease group, which had not been identified previously when analyzing other chlamydial genes. In our further analysis of pmp genes, one of the 19 reference strains, Da/TW-448, appeared to be a recombinant (Gomes et al. 2006). While no clinical strains were analyzed in that study, the finding was surprising as recombination in intracellular pathogens would not be expected based on the organism’s need to reside, while metabolically active, in a cytoplasmic vacuole, which theoretically limits the possibility of genetic exchange between strains.
The only other evidence for recombination in C. trachomatis is confined to a single gene, ompA. Previous examples included visually detected ompA mosaics comprised of two trachoma strains or two urogenital strains (Dean et al. 1992; Brunham et al. 1994; Hayes et al. 1994). More recently, we confirmed three additional ompA mosaics, including D and L1, which involved exchange of an ompA T cell epitope, B and D, which was isolated from the conjunctiva of a trachoma patient, and Ba and D, from the urogenital tract, revealing mutations associated with differential cellular appetence (Millman et al. 2001, 2004). We also found that ompA sequences differed from the expected pmpC sequence, although the breakpoint could not be identified because these two genes are far apart (Gomes et al. 2004).
Recombination has also been reported for the obligate intracellular organisms Rickettsia and Wolbachia. In the human pathogen Rickettsia, genomic analysis revealed infrequent recombination (Jiggins 2006). Wolbachia, which are transmitted by inheritance in arthropods and nematodes, exhibit widespread recombination and insertion sequence (IS)-like elements that suggest a chimeric origin (Baldo et al. 2006). Among species of the Chlamydophila genus, genome analysis suggests genetic exchange as the origin of the add-guaAB gene cluster and copN, which are located in the plasticity zone (PZ), an area of heterogeneity between genera and among species (Read et al. 2003; Thomson et al. 2005). An IS that is homologous to IS605 of Helicobacter pylori has also been discovered within a tetracycline resistance island in Chlamydia suis (Dugan et al. 2004). While these cumulative studies suggest that horizontal gene transfer has almost certainly occurred in Chlamydiaceae, there have been no data capable of addressing whether recombination is as infrequent as speculated or as commonplace as point substitution. Nor has there been a systematic search for recombination between loci across the genome in any Chlamydiaceae species, including C. trachomatis. Here, we sequenced multiple complete loci dispersed throughout the chromosome for the 19 C. trachomatis laboratory-adapted reference strains and 10 present-day ocular and urogenital clinical isolates using phylogenetic reconstructions, genetic matrices, and statistically based recombination programs to determine the extent and significance of recombination.
We provide evidence that interstrain recombination is rampant in this organism and is the dominant source of genetic diversification. Ten of ten clinical isolates are recombinants at multiple loci with no two belonging to the same clonal lineage. We identify two statistically significant hotspots for recombination downstream from ompA, the study of which could eventually allow the mechanism by which C. trachomatis recombinant evolution occurs to be discovered.
Results
Phylogenetic and genomic loci analyses of C. trachomatis reference strains and clinical isolates
Seventeen loci (∼40,000 bp or 4% of the chromosome) were selected for sequencing in the C. trachomatis genome based on three criteria: (1) substantial variation in particular loci that was predicted based on microarray studies of 15 reference strains (Brunelle et al. 2004); (2) intergenic regions adjacent to ompA where mosaic structures have been identified for C. trachomatis strains (Hayes et al. 1994; Millman et al. 2001, 2004); and (3) genes dispersed throughout the chromosome for broad genome coverage. Although well-separated housekeeping genes would have sufficed for detecting the presence of widespread recombination (Feil and Spratt 2001), we anticipated that, by sequencing more polymorphic genes, breakpoints could be identified. We purposely did not select the PZ as this is an area of known variation and is likely involved in highly localized horizontal gene transfer that might have no bearing on evolutionary dynamics occurring more broadly in the genome.
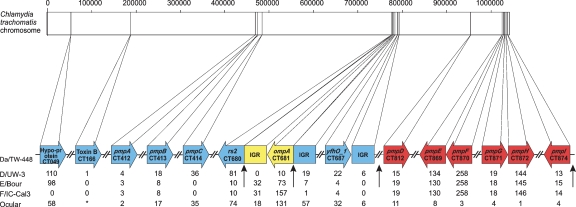
Figure 1 shows the results of analysis of the 17 loci for the 19 reference strains. Phylogenetic trees constructed from the different loci are incongruent (Fig. 2; Supplemental Fig. 1), suggesting recombination. We analyzed the nonsynonymous/synonymous substitution ratios (ω = dN/dS) (Yang and Nielsen 2000) for each chromosome region to identify any distortion in the evolution of a particular locus that might not support recombination as the cause of the difference in tree phylogenies. All values are <1, pointing to an overall evolutionary protein conservation (Supplemental Fig. 2). The %G:C content might indicate insertion of foreign DNA and was calculated for each locus. The content ranged from 38% to 45%, the differences of which are not significant given the standard deviation of nearly 3% for genes in the C. trachomatis genome and a mean of ∼41% (Stephens et al. 1998; Carlson et al. 2005). Contingency matrices (data not shown) also support recombination. These data collectively indicate a mosaic structure for reference strain Da/TW-448.
Figure 1.
Mosaic structure of Da/TW-448. The linear representation of the ∼1.05 Mb chromosome and nucleotide location (vertical lines) of the 17 sequenced loci are shown. Reference strains involved in the Da/TW-448 mosaic are shown in colored boxes: blue (F/IC-Cal-3), yellow (D/UW-3), and red (A/Har-13, B/TW-5, Ba/Apache-2, C/TW-3), below which is the genetic distance between Da/TW-448 and the reference strains. Vertical arrows represent statistically confirmed and putative crossover regions. The chart shows the genetic distance (based on the number of nucleotide differences) between Da/TW-448 and D/UW-3, E/Bour, F/IC-Cal3 and the mean of the ocular reference group (strains A, B, Ba, and C) for each genomic region analyzed. *CT166 (toxin B-related protein) was included as it represents 2865 bp for trachoma strains (except B/TW-5, which does not contain this gene), and 1920 bp for urogenital reference strains.
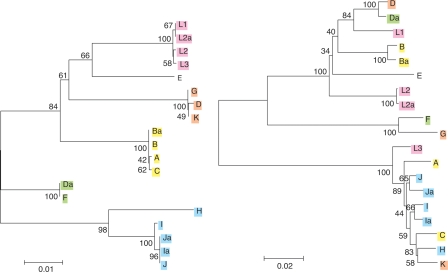
Figure 2.
Phylogenetic reconstructions. Reconstructions of the evolutionary history of (left) CT049 and (right) ompA are based on the respective genomic sequences of C. trachomatis reference strains. Strains are color-coded based on the main lineages of the CT049 tree. Strains responsible for the three different disease groups of C. trachomatis (A, B, Ba, and C for trachoma; L1, L2, L2a, and L3 for invasive STDs; and H, I, J, Ja, I, and Ia for noninvasive STDs and likely persistent infection; Dean et al. 2000) form distinct clusters in the CT049 tree, but not in the ompA tree. They also cluster in trees for CT166 and pmpC (see Supplemental material), indicating the possibility of a clonal frame to which ompA does not belong (see Discussion). The values at the nodes are bootstrap that support levels representing the percentage of 1000 bootstrap resamplings in which the strains to the right form a clade. Amino acid trees (data not shown) are similar to the nucleotide trees.
Guided by the Da/TW-448 structure, we selected seven of the 17 loci that represented four well-separated regions of the chromosome for sequencing the 10 clinical isolates. Trees representing the phylogenetic reconstructions are incongruent for all genomic regions of all clinical isolates (Supplemental Fig. 3). The two least incongruent loci, CT049 and pmpC (Supplemental Fig. 3A and B, respectively), differ only in deeper nodes corresponding to recombination in the evolution of the reference strains, but not more recently in the clinical isolates. Other pairs of loci indicate recombination in the evolution of at least some of the reference strains and all of the clinical isolates. Similar analyses as for the reference strains, including dN/dS (Supplemental Fig. 4), although no dN/dS analysis was performed over specific peptide regions of a protein, %G:C content, and contingency matrices (data not shown), indicate that recombination was the likely explanation for the incongruent trees. These data indicate extensive recombination where each clinical isolate is a distinct mosaic (Fig. 3).
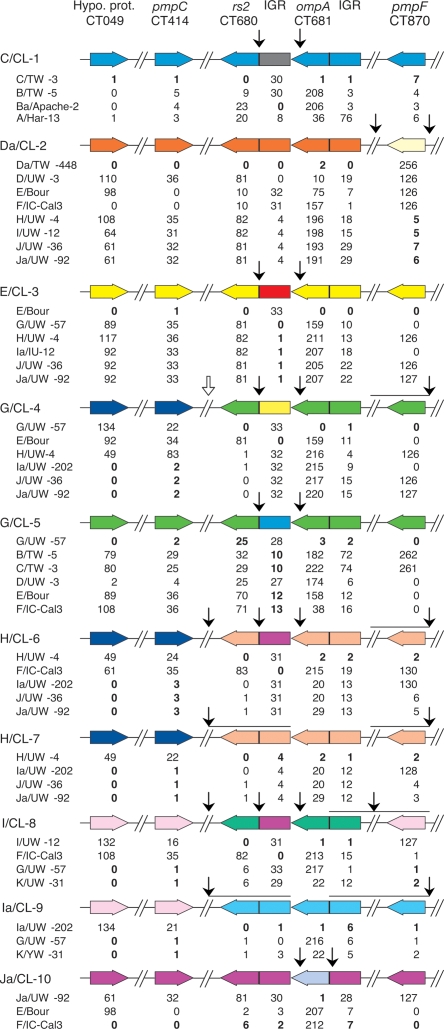
Figure 3.
Linear representation of the proposed chromosomal mosaics for C. trachomatis clinical isolates. The seven loci are represented by colored boxed arrows, which show the coding strand direction of each gene. The vertical arrows represent the deduced crossovers (arrows between noncontiguous sequences) and statistically confirmed hotspots for recombination (arrows between rs2 and the rs2/ompA IGR, and between ompA and the rs2/ompA IGR). The open arrow represents a putative crossover for G/CL-4 as the assignment of rs2 is ambiguous for this isolate. Horizontal lines under arrows that represent deduced crossovers denote the potential area over which the crossover may occur. The chart below each color scheme shows the genetic distance (based on number of nucleotide differences) between the clinical isolate and the reference strain lineages that are likely involved in the mosaic, noted in bold.
Determination of breakpoint regions for Da/TW-448
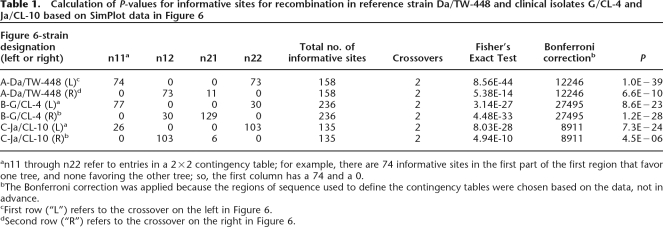
Based on the trees (Supplemental Fig. 1) and contingency matrices (data not shown), the proposed mosaic structure for Da/TW-448 involves four breakpoints (Fig. 1). Figure 4 shows the results of SimPlot analyses, which includes the maximum χ2 test, that was used to determine the regions within which the breakpoints are most likely found. The Recombination Identification Program showed similar results (data not shown). Table 1 shows the calculation of P-values for the informative sites (i.e., sites where Da/TW-448 agrees with one putative donor strain on one side of the breakpoint region and agrees with a different putative donor on the other side; Fig. 4A1) that support each region. The 2×2 matrix for each region is denoted n for informative sites for each topology in each area, from which the Fisher’s Exact Test is calculated. Also shown is the total number of informative sites (t) and the number of breakpoint regions (c) used to analyze the sequence; the Bonferroni correction was used as the number of ways to distribute c in the intervals between the t sites (t − 1 choose c). P-values for the informative sites occurring in a nonrecombinant area areP = 1 × 10−39 for the first region (rs2 and the rs2/ompA IGR) and P = 6.6 × 10−10 for the second (ompA and ompA/pbpB IGR) (Fig. 1; Table 1). Two other breakpoint regions between the yfh0_1/parB IGR and pmpD and between pmpI and CT049 are suggested, but the exact locations could not be determined as we did not sequence the regions between these loci (Fig. 1).
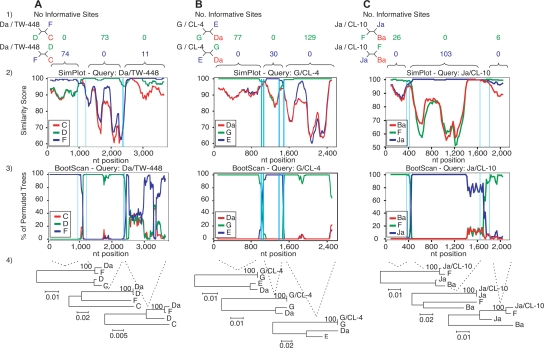
Figure 4.
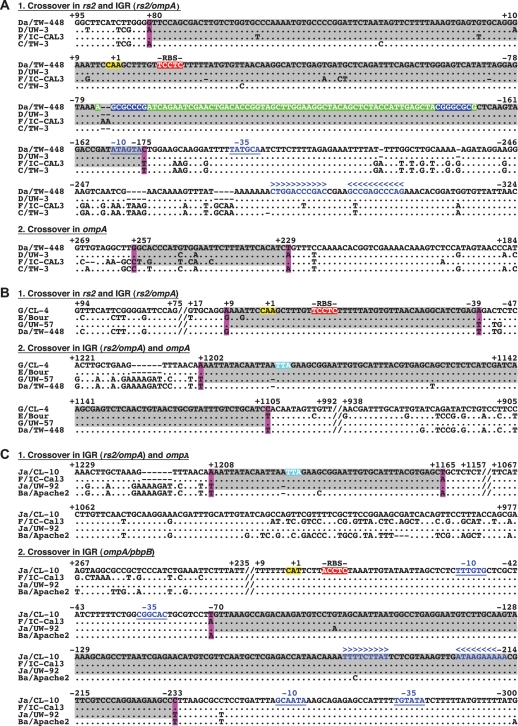
SimPlot representation of the putative crossover regions for the reference strain Da/TW-448 (A) and the clinical isolates G/CL-4 (B) and Ja/CL-10 (C). (1) Number of informative sites shared by the recombinant sequences (black) and the parental reference strains (blue and green). The outgroup sequence is shown in red. Four-member trees consistent with these sites are also shown for each region adjacent to the crossover region. (2) Similarity plot between each recombinant sequence and the respective parental reference strains, with a sliding window size of 200 bp and a step size of 20 bp. (3) BootScan analysis (window size 200 bp; step size 20 bp) showing the phylogenetic relatedness (% of permuted trees) between these sequences. For (2) and (3), the crossover regions are located between each pair of vertical lines, and nucleotides at the bottom of each plot correspond to alignment positions (not to chromosomal locations) of the continuous genomic regions analyzed. For Da/TW-448 and G/CL-4, the genomic region involved the rs2 gene to the ompA/pbpB IGR, while for Ja/CL-10, the region included only the rs2/ompA IGR, ompA, and the ompA/pbpB IGR. (4) Phylogenetic reconstructions for each specific region bounded by the recombination breakpoint region supporting each crossover (1000 bootstrapped trees).
Table 1.
Calculation of P-values for informative sites for recombination in reference strain Da/TW-448 and clinical isolates G/CL-4 and Ja/CL-10 based on SimPlot data in Figure 6
an11 through n22 refer to entries in a 2×2 contingency table; for example, there are 74 informative sites in the first part of the first region that favor one tree, and none favoring the other tree; so, the first column has a 74 and a 0.
bThe Bonferroni correction was applied because the regions of sequence used to define the contingency tables were chosen based on the data, not in advance.
cFirst row (“L”) refers to the crossover on the left in Figure 6.
dSecond row (“R”) refers to the crossover on the right in Figure 6.
Based on these results, Figure 4A2 shows the most likely position of the first breakpoint region that is confined to a 254-bp area, while the second region is restricted to a 29-bp area. These two breakpoint regions are bounded by the outer most informative sites that support one of the three phylogenetic trees shown in Figure 4A4. Figure 5A1, 2 shows the nucleotide sequence of the 254-bp region, which includes a tRNA-Gly-2 with characteristic inverted repeats, a ribosomal binding site (RBS) and promotor sites, and the 29-bp region, respectively.
Figure 5.
Nucleotide sequences of crossovers for reference strain Da/TW-448 (A) and clinical isolates G/CL-4 (B) and Ja/CL-10 (C). Numbers represent positions relative to the start codon of rs2 or ompA (highlighted in yellow). The stop codon of ompA is highlighted in blue. Crossover regions are highlighted in gray bordered by informative sites (fuchsia) from SimPlot/BootScan analysis. The tRNA is highlighted in green with inverted repeats (blue). Putative ribosomal binding sited (RBS) are highlighted in red. An imperfect and a perfect palindrome denote the putative terminators in blue below the symbols >>> <<<. Putative promotor regions are in blue characters and underlined; spacer A/T regions occur downstream from each −35 promoter element.
Determination of breakpoint regions for 10 C. trachomatis clinical isolates
All 10 clinical isolates contain at least two breakpoint regions denoted by black arrows as shown in Figure 3, which are supported by phylogenetic trees (Supplemental Fig. 2) and contingency matrices (data not shown). Interestingly, two pairs of clinical strains with identical ompA sequences (G/CL-4 and G/CL-5; H/CL-6 and H/CL-7) have varying numbers of breakpoint regions composed of DNA descended from different reference strain lineages. G/CL-5 appears to be composed of trachoma strains B and C, which are known to infect the urogenital tract (Millman et al. 2006). Clinical isolate Da/CL-2 is most similar to Da/TW-448, the recombinant nature of which, involving E, F, and D, is again evident (Figs. 1, 3). Also, at the same loci where Da/TW-448 is derived from trachoma strains A, B, Ba, and C, Da/CL-2 is derived from strains H, I, J, and Ja (Fig. 3).
G/CL-4 and Ja/CL-10 are discussed here in more detail as several clinical isolates share breakpoint regions with these strains in the continuous, sequenced region between rs2 and the ompA/pbpB IGR (Fig. 3). Based on SimPlot analyses (Fig. 4), G/CL-4 has two breakpoint regions with P-values of P = 1.2 × 10−28 for the first region (rs2 and rs2/ompA IGR) and P = 8.6 × 10−23 for the second (rs2/ompA IGR and ompA) (Fig. 4B1; Table 1). The most likely positions for the breakpoint regions are within a highly conserved stretch of 48 bp for the first region (Fig. 4B2) in proximity to an RBS, as shown in the nucleotide sequence in Figure 5B1, and within a highly conserved stretch of 98 bp for the second (Figs. 4B2, 5B2). The informative sites support the phylogenetic trees as shown in Figure 4B4.
Figure 4 shows the two breakpoint regions for Ja/CL-10 with P = 7.3 × 10−24 for the first region (ompA and rs2/ompA IGR) and P = 4.5 × 10−6 for the second (ompA/pbpB IGR) (Fig. 4C1; Table 1). The most likely position of the first and second breakpoint regions are within a highly conserved stretch of 44 bp (Figs. 4C2, 5C1) and 164 bp (Figs. 4C2, 5C2), respectively. The latter occurs in proximity to an RBS (Fig. 5C2). These regions are flanked by informative sites that support distinct phylogenetic trees (Fig. 4C4).
Chromosomal hotspots
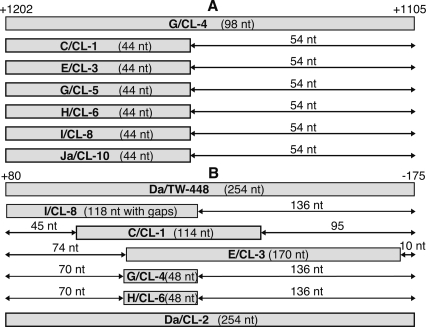
There are a total of 16 identified breakpoints of recombination in the 3.7-kb region of rs2 to the ompA/pbpB IGR (Fig. 3). Two of the 16 breakpoints are not shown in Figure 3; Da/CL-2 is homologous to Da/TW-448 in all loci except for pmpF (Fig. 1), and the regions for G/CL-5 could not be precisely localized as the tree strongly suggests that the donor strain is not represented among the strains (Supplemental Fig. 2D). The 16 breakpoints form two clusters. The first cluster encompasses the rs2/ompA IGR and 3′ terminus of ompA where six clinical isolates (C/CL-1, E/CL-3, G/CL-5, H/CL6, I/CL-8, and Ja/CL-10) have the same 44-bp breakpoint region, the alignment of which is depicted in Figure 6A. The second cluster encompasses the 5′ end of rs2 and the rs2/ompA IGR where seven clinical isolates (C/CL-1, Da/CL-2, E/CL-3, G/CL-4, G/CL-5, H/CL-6, and I/CL-8) are restricted to different bp segments, but all except G/CL-5 are confined to a 254-bp span and overlap a single 48-bp breakpoint region. The alignment of these strains is shown in Figure 6B.
Figure 6.
Representation of the nucleotide positions of the two chromosomal hotspots. Informative sites bound the recombination breakpoints to within the shaded box for each isolate. (A) The 44-bp hotspot overlaps the rs2/ompA IGR and ompA and is within the 98-bp crossover region of clinical isolate G/CL-4. (B) The 48-bp hotspot involves the rs2 and the rs2/ompA IGR and is within the 254-bp crossover region of reference strain Da/TW-448. For A and B, the numbers +1202 and +1105, and +80 and −175 are the genomic positions relative to the start codon of ompA and rs2, respectively.
Analysis of the genotype assignments in Figure 3 reveals that no two of the recombination events in this region involve the same combinations of genotypes. Moreover, the phylogenies (Supplemental Fig. 3) show that the corresponding horizontal transfers are between pairs of branches in the tree that are distinguished by reasonable (>60%) bootstrap support in at least one tree. We conclude that no two recombinations in this region are descended from a single ancestral event and can be treated as statistically independent. In fact, only one pair of events in clinical samples H/CL-6 and H/CL-7 plausibly shares any recombinant events since the time of divergence of the reference strains (Fig. 3).
To compute P-values for the two clusters, we considered a null hypothesis that the 16 events are distributed randomly over this continuous 3.7-kb sequence. However, the events within ∼200–300 bases of either end might not have enough informative sites to be confidently assigned. Considering this, we estimated the probability of the observed six or more sequenced breakpoint regions falling in the 44-bp region, if there was no hotspot, to be P = 2.9 × 10−6, after applying a multiple-test correction. Using a conservative window of 254 bp for the second breakpoint region, the probability for the observed six or more of the sequenced crossovers falling within the window around the 48-bp region was estimated to be P = 0.011 after applying a multiple-test correction (see Methods).
An analysis considering only variable sites resulted in even more significant P-values, indicating that the appearance of hotspots is not caused by the nonuniform distribution of variable sites (data not shown).
Discussion
While recombination has been widely documented for extracellular or facultative intracellular organisms such as the genera Neisseria (Gibbs et al. 1989), Borrelia (Cadavid et al. 1994), Leptospira (Haake et al. 2004), and Anaplasma (Brayton et al. 2002), reports for obligate intracellular organisms are rare (Ogata et al. 2005; Baldo et al. 2006; Jiggins 2006). Our work provides the first extensive recombination study in Chlamydiaceae and is the first comprehensive recombination analysis performed in a human obligate intracellular pathogen. Even with the complete genomes of nine Chlamydiales species and strains, including two C. trachomatis reference strains (but no recent clinical isolates), it was not possible to predict the rampant recombination we found. The widespread recombination occurring in C. trachomatis implies that the combinations of genes that are likely to explain disease phenotype are not, for the most part, all genetically linked. Our discovery has major biomedical implications and demonstrates the importance in this organism of intensive sampling (multiple clinical isolates and reference strains) at multiple loci.
Genetic recombination is the only plausible mechanism of generation of the reference strain Da/TW-448 and the 10 clinical mosaic strains, as the accumulation of point mutations was statistically highly improbable. In fact, 14 of the 16 breakpoint regions are located in one of the two clusters (Fig. 6), making it unlikely that the crossovers occurred at random. Consequently, we consider the 48-bp and 44-bp regions to be hotspots for interstrain recombination.
The observed pattern of mosaicism indicates statistically significant hotspots for recombination, which implies either a site-specific recombination mechanism or selection for the resulting recombinant genomes. However, the fact that the data are consistent with many breakpoint regions occurring at exactly the same location suggests the former, possibly in combination with the latter. The regions lack recognizable sequences such as Chi sites or direct target repeats that are associated with some of the best-studied mechanisms of gene mobility and rearrangement (Gomes et al. 2004; Ogata et al. 2005; Baldo et al. 2006). However, Chi sites differ across bacteria and have not yet been determined for Chlamydiaceae. Hotspots adjacent to important genomic loci, notably ompA, and its neighboring IGRs, which contain regulatory regions (Fig. 5), may indicate that gene swapping via recombination is not random. Indeed, one hotspot contains a tRNA gene. tRNAs are often associated with pathogenicity islands and have recently been shown to be hotspots for horizontal gene transfer among extracellular bacteria (Bishop et al. 2005). Natural selection may have favored evolutionary mechanisms that allow lateral gene transfer between coinfecting members of the species near conserved regions, such as RBSs, where function would not be disturbed, resulting in the genesis of successful new clones, as appears to be the case in this study.
While recombination would seem unlikely for an obligate intracellular pathogen that is sequestered from other strains by inhabiting an intracytoplasmic vacuole, various types of genetic exchange would be consistent with the knowledge that DNA repair and recombination systems, including recBCD and xer enzymes, are well represented in the C. trachomatis genome (Supplemental Table 1). Simultaneous infections of a single host cell with two C. trachomatis strains in which fusion of the respective vacuoles occurs have been documented in in vitro systems (Ridderhof and Barnes 1989), and mixed infections have been reported in vivo (Brunham et al. 1994; Hayes et al. 1994; Dean et al. 2000). Yet, this raises the question of whether there are specific mechanisms that bring coinfecting chlamydiae into direct contact within the vacuole to facilitate gene transfer. The IS in C. suis (Dugan et al. 2004), the evidence for horizontal gene transfer involving the add-guaAB gene cluster and copN of Chlamydophila species (Read et al. 2003; Thomson et al. 2005), and clustering of pmps in each Chlamydiaceae chromosome (Grimwood and Stephens 1999) along with gene duplication in different coding directions suggest recombinant events involving the various genomes. These cumulative data, then, support the unexpected recombinant and divergent clonal lineages we discovered for Da/TW-448 and the clinical isolates of C. trachomatis.
Our study shows that ompA is not genetically linked with a great deal of the genome, including the PZ region that contains a tryptophan synthase that may correlate with tissue tropism (Caldwell et al. 2003). We found no evidence for recombination in the chromosome region between CT049 and CT414 (Figs. 1, 2) that would indicate horizontal gene transfer including the PZ (∼CT152–CT176), but our study would not have detected exchange of excised PZs. Our findings of recombination throughout the genome will be essential for interpretation of any future studies that attempt to track PZ exchange across clinical isolates.
Traditional C. trachomatis classification based on polymorphisms in one or two genes appears to be too restrictive since the similarity of the clinical isolates with the corresponding reference strain is sometimes confined to only a single gene. This is the case for Ja/CL-10, where only the ompA gene matches genovariant Ja/UW-92 (Fig. 3), and is further supported by the fact that all circulating clinical isolates in this study reveal a different mosaic structure, even for those with identical ompA sequences (e.g., G/CL-4 and G/Cl-5, and H/CL-6 and H/CL-7) (Fig. 3). The contradictory results on the association between serotype or ompA-genotype and clinical findings reported in the literature (Workowski et al. 1994; Morré et al. 2000; Anttila et al. 2001; Millman et al. 2006) are to be expected given our results. Indeed, our data suggest that there are many more divergent clones than previously imagined for C. trachomatis. In light of our findings, strain typing should involve a number of loci combined in multilocus sequence-based typing (MLST) strategies, which have been successfully used for typing disease- and nondisease-causing lineages among 21 extracellular bacteria and yeast to date (www.mlst.net) (Maiden et al. 1998; Enright et al. 2001; Feil et al. 2003).
A broader study may reveal the time scale in which a given clonal strain persists in disease populations. Considering that our results come from a survey involving ∼40,000 bp of each ∼1.05 Mb chromosome, our findings likely represent the “tip of the iceberg” of a still more complex picture. Significantly, when linkage disequilibrium has been observed between alleles for C. trachomatis, which might suggest a lack of recombination, the analysis was based on only reference strains and partial sequences of a few genes in close proximity to one another (Stothard et al. 2003). By selecting seven loci dispersed throughout the genome for all reference strains and 10 recent clinical isolates, we were able to detect the emergence of mosaic strains that are likely occurring from extensive recombination more recently in time. Although the clinical mosaics presented in this study are graphically represented as chimeras of parental reference strains, in reality, recombination occurs between contemporary circulating strains. The implications of these findings are that contemporary human chlamydial diseases are not caused by clonal descendents of reference strains but by strains evolved from multiple recombinations of ancestral lineages.
The degree of widespread recombination we describe far exceeds that for Rickettsia, the only other intracellular human pathogen in which recombination is known to occur. We speculate that comparative genomics of C. trachomatis reference strains and additional clinical isolates would readily allow the identification of other recombination sites and possibly the mechanism(s) for each. Based on our data (Figs. 1, 3), a clonal frame (Milkman and Bridges 1990; Smith et al. 1993), in which clinical genotypes consistently agree, may exist from CT049 to CT414 (∼400 kb apart), possibly extending in either direction. However, phylogenies for the reference strains at these two loci are not concordant, implying that recombination takes place in this region at a slower rate than in the vicinity of ompA. The clonal frame appears to degrade at pmpF and disappears between rs2 and ompA. According to the positions of the putative breakpoints (denoted by vertical arrows in Fig. 3), transferred segments appear to involve the IGR downstream from ompA (∼350 bp), ompA (∼1200 bp), and rs2 (∼900 bp). The segments are primarily under 10 kb, suggesting natural DNA transformation as a reasonable mechanism for genetic exchange, which is consistent with what has been reported for over 44 bacteria (Lorenz and Wackernagel 1994).
Recombination provides a mechanism where genes encoding polymorphic antigens, pathogenic factors, or proteins involved in cell appetence could hypothetically be transferred and confer biological advantages for the chlamydial receptor strain. Thus, our results do not point to specific events that are responsible for different pathogenicities but, instead, suggest a new way to dissect the genetic basis for the disease pathology of clinical strains. Our current understanding of C. trachomatis will have to be revised as present-day clinical isolates are used in in vitro and in vivo research studies in tandem with reference strains to decipher the molecular and genetic mechanisms involved in contemporary and emerging human chlamydial diseases. In addition, our findings suggest the need for an expansive genomic exploration of clonal populations of C. trachomatis as well as of other obligate intracellular pathogens that may also be undergoing extreme recombination.
Methods
C. trachomatis reference strains and clinical isolates
The 19 reference strains (A/Har-13, B/TW-5, Ba/Apache-2, C/TW-3, D/UW-3, Da/TW-448, E/Bour, F/IC-Cal3, G/UW-57, H/UW-4, I/UW-12, Ia/IU-4168, J/UW-36, K/UW-31, L1/440, L2/434, L2a/TW-396, and L3/404), including genovariant Ja/UW-92 and 10 clinical isolates representing eight ompA genotypes (C, Da, E, 2 G, 2 H, I, Ia, and Ja) from women with trachoma or urogenital infections were analyzed. We designated each clinical isolate in alphabetical order as C/CL-1, Da/CL-2, etc.
Cell culture of all C. trachomatis strains was performed as previously described (Dean et al. 2000; Dean and Powers 2001). After Renografin purification, genomic DNA was extracted using High Pure PCR Template Preparation Kit (Roche Diagnostics) according to manufacturer’s instructions. Confirmation of reference strains and identification of clinical isolates were performed using ompA genotyping with BLAST comparison of the sequences in GenBank as previously described (Gomes et al. 2004).
Sequencing of genomic regions
Approximately 40,000 bp of the ∼1.05 Mb C. trachomatis chromosome was analyzed for each reference strain. PCR primers (Supplemental Table 2) were designed using Primer Select software (DNASTAR) based on published genome sequences of reference strains D/UW-3 and A/Har-13 (GenBank AE001273 and CP000051, respectively). Primers for amplification and sequencing of pmp genes were used as previously described (Gomes et al. 2006). For CT166, only Da/TW-448 was sequenced, as sequences for other reference strains are available in GenBank. PCR reagents and thermocycling profiles were as previously described (Gomes et al. 2006), except for annealing temperatures and extension times: 54°C and 2 min, 15 sec, for CT049-1/CT049-2; 55°C and 1 min, 40 sec, for rs2-1/rs2-2 and rs2-1/rs2-3; 56°C and 1 min, 30 sec, for pbpB-1/pbpB-2; 56°C and 2 min for yfh0-1/yfh0-2. Amplicons were visualized in ethidium bromide-stained 0.8% agarose gels, purified, and sequenced as previously described (Gomes et al. 2006).
Determination of %G:C content
The %G:C content for each loci was determined using EditSeq software (DNASTAR). We also searched for direct target repeats and Escherichia coli-like Chi sites that may indicate regions of recombination using multiple Web-based freeware, including BLAST, and search engines.
Analysis of the molecular evolution of genomic regions
The molecular evolution of the loci was evaluated using the Nei-Gojobori method (Nei and Kumar 2000) to estimate the ratio of dN/dS substitutions. These values were normalized (P-distance model) against the number of potential dN and dS sites because the latter is much smaller than the former. Significant differences in mean dN and dS were determined by comparing 95% confidence intervals (CI). An SE computation was performed by bootstrap analysis with 1000 replications.
Phylogenetic and statistical analyses
Phylogenetic data were generated using MEGA (http://www.megasoftware.net) as previously described (Gomes et al. 2004) where Neighbor-joining trees (Saitou and Nei 1987; Nei and Kumar 2000) were created using the Kimura-2-parameter model (Kimura 1980) that takes into account substitution rates while assuming that the four nucleotide frequencies are the same and that rates of substitution do not vary among sites (Nei and Kumar 2000). Phylogenetic reconstructions were also performed at the protein level using the Gamma distance model that takes into account the dissimilarity of the substitution rates among sites (Nei and Kumar 2000). Additionally, one matrix was generated for each of the loci that were analyzed for reference strains and clinical isolates. The numbers represent absolute nucleotide differences between each pair of strains under comparison for the total number of valid sites.
Determination of breakpoint regions
A precise search for potential mosaic structures was performed using SimPlot (http://sray.med.som.jhmi.edu/SCRoftware/) (Lole et al. 1999) to calculate and plot the similarity of one putative recombinant sequence (Query) to the other sequences in a sliding window size of 200 bp moved across the alignment in a step size of 20 bp. Nucleotide pairwise distances were calculated using the Kimura-2-parameter method (gaps excluded; ts/tv of 2.0). A Bootscanning analysis (Salminen et al. 1995) was also performed for Da/TW-448 and all clinical isolates. Each was compared with sequences from the probable parental strain(s) and a known outgroup sequence. At each nucleotide window range, a phylogenetic analysis was performed using the Neighbor-joining topology on the basis of pairwise genetic distances (Kimura-2-parameter method; gaps strip on; ts/tv of 2.0). Bootstrap confidence levels were determined by 100 replicates. Significant changes in phylogenetic relationships from window to window resulted in changes in BootScan values that were indicative of probable recombination events. SimPlot was also used to identify informative sites (Robertson et al. 1995) where two sequences share one specific nucleotide but the other two share a different nucleotide. Each informative site supports one of the three possible phylogenetic relationships among the four taxa. The likelihood that the observed distribution of sites favoring specific phylogenetic groupings might occur randomly was assessed using the maximum χ2 test. The most likely crossover region occurred where the observed distribution is least likely to occur randomly (maximum χ2 value).
The null hypothesis for recombination was that informative sites are drawn from a single distribution. A P-value for any specified breakpoint is given by Fisher’s Exact Test. Because breakpoints are not specified in advance, we applied a Bonferroni multiple test factor for the number of ways to choose breakpoints. This makes the test conservative because the different hypotheses corresponding to different breakpoints are highly correlated. Some choices of breakpoints will have very little power, further reducing the effective number of tests. A more complex calculation that avoids our conservative approximations is given by Halpern (2000).
To support the recombination regions given by SimPlot and to accurately infer the phylogenetic relationships of the putative recombinant strains, the defined breakpoint regions were used to divide the alignments into delimited genomic regions, and phylogenetic trees were inferred by Neighbor-joining (Kimura-2-parameter) analysis.
For comparative purposes with data obtained using SimPlot, we also used the RIP (http://hiv-web.lanl.gov/) (Smith 1992).
Statistical analyses of chromosomal hotspots
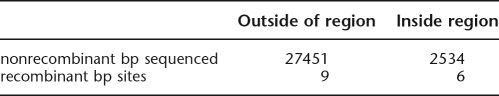
We considered conservative cases with equal-sized windows for the calculations. For the 44-bp breakpoint region, which is shared by seven clinical isolates and termed a cluster, we ignored the seventh event (G/CL-4 with 98 bp) and considered whether six crossovers restricted to 44 bp are overrepresented relative to nine events outside of that region. We framed this as a contingency table with ∼30,000 bp of sequence in each of the 10 clinical isolates, which gives a P = 4.3 × 10−8 (Fisher’s Exact Test):
In this case, the test is not exact because the number of breakpoint regions detected in our 10 clinical sequences cannot exceed 10 in a given window bounded by informative sites, making our result more conservative. To this value, we apply a multiple-test correction (roughly a factor of 3000/44), because the 44-bp region was not specified in advance, resulting in P = 2.9 × 10−6. For the other cluster of 254 bp for seven clinical isolates, we have six events plus a seventh (G/CL-5) that is ignored because it could be outside that region. Our contingency table gives P = 9.4 × 10−4. Corrected by 3000/254, this gives a P = 0.011.
Acknowledgments
We thank Tim Read for critical review of the manuscript and Bette Korber for helpful comments and encouragement along the way. This research was supported by Fundação Para a Ciência e Tecnologia and FEDER (POCTI/39822/MGI/2001) (MJB) and Public Health Service Grants from the National Institutes of Health, R01-AI59647 (DD) and R01-AI39499 (DD).
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession numbers: AY884090–AY884108 (for pmpA), AY884109–AY884127 (for pmpB), AY299408–AY299426 (for pmpD), AY967735–AY967738 (for pmpE), AY887644–AY887662 and DQ065739– DQ065748 (for pmpF), AY967739–AY967757 (for pmpG), AY967759–AY967761 (for pmpH), AY299427–AY299445 (for pmpI), DQ065736–DQ065738, DQ062749–DQ062755, and DQ076723–DQ076741 (for ORF CT049), DQ113596–DQ113614 and DQ076742–DQ076751 (for IGR rs2/ompA), DQ116393–DQ116402 (for ompA), DQ113625– DQ113643 and DQ113615–DQ113624 (for IGR [ompA/pbpB]), DQ151840 (for ORF CT166), DQ151841–DQ151847 and DQ239937–DQ239957 (for rs2), DQ151848–DQ151854 (for yfh0_1 and IGR [yfh0_1/parB]). Clinical isolates have the following designation in GenBank: C/CL-1 (CS-362-07), Da/CL-2 (CS-431/04), E/CL-3 (I-174), G/CL-4 (CS-490/95), G/CL-5 (I-149), H/CL-6 (CS-121/96), H/CL-7 (I-139), I/CL-8 (I-24), Ia/CL-9 (CS-190/96), and Ja/CL-10 (S-91).]
Article published online before print. Article and publication data are at http://www.genome.org/cgi/doi/10.1101/gr.5674706
References
- Anttila T., Saikku P., Koskela P., Bloigu A., Dillner J., Ikaheimo I., Jellum E., Lehtinen M., Lenner P., Hakulinen T., Saikku P., Koskela P., Bloigu A., Dillner J., Ikaheimo I., Jellum E., Lehtinen M., Lenner P., Hakulinen T., Koskela P., Bloigu A., Dillner J., Ikaheimo I., Jellum E., Lehtinen M., Lenner P., Hakulinen T., Bloigu A., Dillner J., Ikaheimo I., Jellum E., Lehtinen M., Lenner P., Hakulinen T., Dillner J., Ikaheimo I., Jellum E., Lehtinen M., Lenner P., Hakulinen T., Ikaheimo I., Jellum E., Lehtinen M., Lenner P., Hakulinen T., Jellum E., Lehtinen M., Lenner P., Hakulinen T., Lehtinen M., Lenner P., Hakulinen T., Lenner P., Hakulinen T., Hakulinen T., et al. Serotypes of Chlamydia trachomatis and risk for development of cervical squamous cell carcinoma. JAMA. 2001;285:47–51. doi: 10.1001/jama.285.1.47. [DOI] [PubMed] [Google Scholar]
- Baldo L., Bordenstein S., Wernegreen J.J., Werren J.H., Bordenstein S., Wernegreen J.J., Werren J.H., Wernegreen J.J., Werren J.H., Werren J.H. Widespread recombination throughout Wolbachia genomes. Mol. Biol. Evol. 2006;23:437–449. doi: 10.1093/molbev/msj049. [DOI] [PubMed] [Google Scholar]
- Bishop A.L., Baker S., Jenks S., Fookes M., Gaora P.O., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Baker S., Jenks S., Fookes M., Gaora P.O., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Jenks S., Fookes M., Gaora P.O., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Fookes M., Gaora P.O., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Gaora P.O., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Anjum M., Farrar J., Hien T.T., Ivens A., Farrar J., Hien T.T., Ivens A., Hien T.T., Ivens A., Ivens A., et al. Analysis of the hypervariable region of the Salmonella enterica genome associated with tRNA(leuX) J. Bacteriol. 2005;187:2469–2482. doi: 10.1128/JB.187.7.2469-2482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert J.F., Koutsky L.A., Suchland R.J., Stamm W.E., Koutsky L.A., Suchland R.J., Stamm W.E., Suchland R.J., Stamm W.E., Stamm W.E. Clinical features of Chlamydia trachomatis rectal infection by serovar among homosexually active men. Sex. Transm. Dis. 1999;26:392–398. doi: 10.1097/00007435-199908000-00006. [DOI] [PubMed] [Google Scholar]
- Brayton K.A., Palmer G.H., Lundgren A., Yi J., Barbet A.F., Palmer G.H., Lundgren A., Yi J., Barbet A.F., Lundgren A., Yi J., Barbet A.F., Yi J., Barbet A.F., Barbet A.F. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol. Microbiol. 2002;43:1151–1159. doi: 10.1046/j.1365-2958.2002.02792.x. [DOI] [PubMed] [Google Scholar]
- Brunelle B.W., Nicholson T.L., Stephens R.S., Nicholson T.L., Stephens R.S., Stephens R.S. Microarray-based genomic surveying of gene polymorphisms in Chlamydia trachomatis. Genome Biol. 2004;5:R42. doi: 10.1186/gb-2004-5-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunham R., Yang C., Maclean I., Kimani J., Maitha G., Plummer F., Yang C., Maclean I., Kimani J., Maitha G., Plummer F., Maclean I., Kimani J., Maitha G., Plummer F., Kimani J., Maitha G., Plummer F., Maitha G., Plummer F., Plummer F. Chlamydia trachomatis from individuals in a sexually transmitted diseases core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J. Clin. Invest. 1994;94:458–463. doi: 10.1172/JCI117347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadavid D., Thomas D.D., Crawley R., Barbour A.G., Thomas D.D., Crawley R., Barbour A.G., Crawley R., Barbour A.G., Barbour A.G. Variability of a bacterial surface protein and disease expression in a possible mouse model of systemic Lyme borreliosis. J. Exp. Med. 1994;179:631–642. doi: 10.1084/jem.179.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell H.D., Wood H., Crane D., Bailey R., Jones R.B., Mabey D., Maclean I., Mohammed Z., Peeling R., Roshick C., Wood H., Crane D., Bailey R., Jones R.B., Mabey D., Maclean I., Mohammed Z., Peeling R., Roshick C., Crane D., Bailey R., Jones R.B., Mabey D., Maclean I., Mohammed Z., Peeling R., Roshick C., Bailey R., Jones R.B., Mabey D., Maclean I., Mohammed Z., Peeling R., Roshick C., Jones R.B., Mabey D., Maclean I., Mohammed Z., Peeling R., Roshick C., Mabey D., Maclean I., Mohammed Z., Peeling R., Roshick C., Maclean I., Mohammed Z., Peeling R., Roshick C., Mohammed Z., Peeling R., Roshick C., Peeling R., Roshick C., Roshick C., et al. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Invest. 2003;111:1757–1769. doi: 10.1172/JCI17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J.H., Hughes S., Hogan D., Cieplak G., Sturdevant D.E., McClarty G., Caldwell H.D., Belland R.J., Hughes S., Hogan D., Cieplak G., Sturdevant D.E., McClarty G., Caldwell H.D., Belland R.J., Hogan D., Cieplak G., Sturdevant D.E., McClarty G., Caldwell H.D., Belland R.J., Cieplak G., Sturdevant D.E., McClarty G., Caldwell H.D., Belland R.J., Sturdevant D.E., McClarty G., Caldwell H.D., Belland R.J., McClarty G., Caldwell H.D., Belland R.J., Caldwell H.D., Belland R.J., Belland R.J. Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect. Immun. 2004;72:7063–7072. doi: 10.1128/IAI.72.12.7063-7072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J.H., Porcella S.F., McClarty G., Caldwell H.D., Porcella S.F., McClarty G., Caldwell H.D., McClarty G., Caldwell H.D., Caldwell H.D. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect. Immun. 2005;73:6407–6418. doi: 10.1128/IAI.73.10.6407-6418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control Prevention, Tracking the hidden epidemics: Trends in STDs in the United States. Department of Health and Human Services; Atlanta, GA: 2005. [Google Scholar]
- Dean D. Pathogenesis of chlamydial ocular infections. In: Tasman W., Jaeger E.A., Jaeger E.A., editors. Duane’s foundations of clinical ophthalmology. Lippincott Williams & Wilkins; Philadelphia, PA: 2002. pp. 1–22. [Google Scholar]
- Dean D., Millman K., Millman K. Molecular and mutation trends analyses of omp1 alleles for serovar E of Chlamydia trachomatis. Implications for the immunopathogenesis of disease. J. Clin. Invest. 1997;99:475–483. doi: 10.1172/JCI119182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D., Powers V.C., Powers V.C. Persistent Chlamydia trachomatis infections resist apoptotic stimuli. Infect. Immun. 2001;69:2442–2447. doi: 10.1128/IAI.69.4.2442-2447.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D., Schachter J., Dawson C.R., Stephens R.S., Schachter J., Dawson C.R., Stephens R.S., Dawson C.R., Stephens R.S., Stephens R.S. Comparison of the major outer membrane protein variant sequence regions of B/Ba isolates: A molecular epidemiologic approach to Chlamydia trachomatis infections. J. Infect. Dis. 1992;166:383–392. doi: 10.1093/infdis/166.2.383. [DOI] [PubMed] [Google Scholar]
- Dean D., Suchland R., Stamm W., Suchland R., Stamm W., Stamm W. Evidence for long-term cervical persistence of Chlamydia trachomatis by omp1 genotyping. J. Infect. Dis. 2000;182:909–916. doi: 10.1086/315778. [DOI] [PubMed] [Google Scholar]
- Dugan J., Rockey D.D., Jones L., Andersen A.A., Rockey D.D., Jones L., Andersen A.A., Jones L., Andersen A.A., Andersen A.A. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob. Agents Chemother. 2004;48:3989–3995. doi: 10.1128/AAC.48.10.3989-3995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright M.C., Spratt B.G., Kalia A., Cross J.H., Bessen D.E., Spratt B.G., Kalia A., Cross J.H., Bessen D.E., Kalia A., Cross J.H., Bessen D.E., Cross J.H., Bessen D.E., Bessen D.E. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 2001;69:2416–2427. doi: 10.1128/IAI.69.4.2416-2427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil E.J., Spratt B.G., Spratt B.G. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 2001;55:561–590. doi: 10.1146/annurev.micro.55.1.561. [DOI] [PubMed] [Google Scholar]
- Feil E.J., Cooper J.E., Grundmann H., Robinson D.A., Enright M.C., Berendt T., Peacock S.J., Smith J.M., Murphy M., Spratt B.G., Cooper J.E., Grundmann H., Robinson D.A., Enright M.C., Berendt T., Peacock S.J., Smith J.M., Murphy M., Spratt B.G., Grundmann H., Robinson D.A., Enright M.C., Berendt T., Peacock S.J., Smith J.M., Murphy M., Spratt B.G., Robinson D.A., Enright M.C., Berendt T., Peacock S.J., Smith J.M., Murphy M., Spratt B.G., Enright M.C., Berendt T., Peacock S.J., Smith J.M., Murphy M., Spratt B.G., Berendt T., Peacock S.J., Smith J.M., Murphy M., Spratt B.G., Peacock S.J., Smith J.M., Murphy M., Spratt B.G., Smith J.M., Murphy M., Spratt B.G., Murphy M., Spratt B.G., Spratt B.G., et al. How clonal is Staphylococcus aureus? J. Bacteriol. 2003;185:3307–3316. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick K.D., Basilion E.V., Hanson C.L., Colchero M.A., Basilion E.V., Hanson C.L., Colchero M.A., Hanson C.L., Colchero M.A., Colchero M.A. Estimating the burden and economic impact of trachomatous visual loss. Ophthalmic Epidemiol. 2003;10:121–132. doi: 10.1076/opep.10.2.121.13899. [DOI] [PubMed] [Google Scholar]
- Gibbs C.P., Reimann B.Y., Schultz E., Kaufmann A., Haas R., Meyer T.F., Reimann B.Y., Schultz E., Kaufmann A., Haas R., Meyer T.F., Schultz E., Kaufmann A., Haas R., Meyer T.F., Kaufmann A., Haas R., Meyer T.F., Haas R., Meyer T.F., Meyer T.F. Reassortment of pilin genes in Neisseria gonorrhoeae occurs by two distinct mechanisms. Nature. 1989;338:651–652. doi: 10.1038/338651a0. [DOI] [PubMed] [Google Scholar]
- Gomes J.P., Bruno W.J., Borrego M.J., Dean D., Bruno W.J., Borrego M.J., Dean D., Borrego M.J., Dean D., Dean D. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J. Bacteriol. 2004;186:4295–4306. doi: 10.1128/JB.186.13.4295-4306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes J.P., Nunes A., Bruno W.J., Borrego M.J., Florindo C., Dean D., Nunes A., Bruno W.J., Borrego M.J., Florindo C., Dean D., Bruno W.J., Borrego M.J., Florindo C., Dean D., Borrego M.J., Florindo C., Dean D., Florindo C., Dean D., Dean D. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: Evidence for serovar da recombination and correlation with tissue tropism. J. Bacteriol. 2006;188:275–286. doi: 10.1128/JB.188.1.275-286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwood J., Stephens R.S., Stephens R.S. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb. Comp. Genomics. 1999;4:187–201. doi: 10.1089/omi.1.1999.4.187. [DOI] [PubMed] [Google Scholar]
- Haake D.A., Suchard M.A., Kelley M.M., Dundoo M., Alt D.P., Zuerner R.L., Suchard M.A., Kelley M.M., Dundoo M., Alt D.P., Zuerner R.L., Kelley M.M., Dundoo M., Alt D.P., Zuerner R.L., Dundoo M., Alt D.P., Zuerner R.L., Alt D.P., Zuerner R.L., Zuerner R.L. Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J. Bacteriol. 2004;186:2818–2828. doi: 10.1128/JB.186.9.2818-2828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern A.L. Multiple-changepoint testing for an alternating segments model of a binary sequence. Biometrics. 2000;56:903–908. doi: 10.1111/j.0006-341x.2000.00903.x. [DOI] [PubMed] [Google Scholar]
- Hayes L.J., Yearsley P., Treharne J.D., Ballard R.A., Fehler G.H., Ward M.E., Yearsley P., Treharne J.D., Ballard R.A., Fehler G.H., Ward M.E., Treharne J.D., Ballard R.A., Fehler G.H., Ward M.E., Ballard R.A., Fehler G.H., Ward M.E., Fehler G.H., Ward M.E., Ward M.E. Evidence for naturally occurring recombination in the gene encoding the major outer membrane protein of lymphogranuloma venereum isolates of Chlamydia trachomatis. Infect. Immun. 1994;62:5659–5663. doi: 10.1128/iai.62.12.5659-5663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins F.M. Adaptive evolution and recombination of Rickettsia antigens. J. Mol. Evol. 2006;62:99–110. doi: 10.1007/s00239-005-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Lavreys L., Chohan V., Overbaugh J., Hassan W., McClelland R.S., Kreiss J., Mandaliya K., Ndinya-Achola J., Baeten J.M., Chohan V., Overbaugh J., Hassan W., McClelland R.S., Kreiss J., Mandaliya K., Ndinya-Achola J., Baeten J.M., Overbaugh J., Hassan W., McClelland R.S., Kreiss J., Mandaliya K., Ndinya-Achola J., Baeten J.M., Hassan W., McClelland R.S., Kreiss J., Mandaliya K., Ndinya-Achola J., Baeten J.M., McClelland R.S., Kreiss J., Mandaliya K., Ndinya-Achola J., Baeten J.M., Kreiss J., Mandaliya K., Ndinya-Achola J., Baeten J.M., Mandaliya K., Ndinya-Achola J., Baeten J.M., Ndinya-Achola J., Baeten J.M., Baeten J.M. Hormonal contraception and risk of cervical infections among HIV-1-seropositive Kenyan women. AIDS. 2004;18:2179–2184. doi: 10.1097/00002030-200411050-00010. [DOI] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C., Ingersoll R., Sheppard H.W., Ray S.C., Sheppard H.W., Ray S.C., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M.G., Wackernagel W., Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden M.C., Bygraves J.A., Feil E., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Bygraves J.A., Feil E., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Feil E., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Zhang Q., Zhou J., Zurth K., Caugant D.A., Zhou J., Zurth K., Caugant D.A., Zurth K., Caugant D.A., Caugant D.A., et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkman R., Bridges M.M., Bridges M.M. Molecular evolution of the Escherichia coli chromosome. III. Clonal frames. Genetics. 1990;126:505–517. doi: 10.1093/genetics/126.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman K.L., Tavare S., Dean D., Tavare S., Dean D., Dean D. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J. Bacteriol. 2001;183:5997–6008. doi: 10.1128/JB.183.20.5997-6008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman K., Black C.M., Johnson R.E., Stamm W.E., Jones R.B., Hook E.W., Martin D.H., Bolan G., Tavaré S., Dean D., Black C.M., Johnson R.E., Stamm W.E., Jones R.B., Hook E.W., Martin D.H., Bolan G., Tavaré S., Dean D., Johnson R.E., Stamm W.E., Jones R.B., Hook E.W., Martin D.H., Bolan G., Tavaré S., Dean D., Stamm W.E., Jones R.B., Hook E.W., Martin D.H., Bolan G., Tavaré S., Dean D., Jones R.B., Hook E.W., Martin D.H., Bolan G., Tavaré S., Dean D., Hook E.W., Martin D.H., Bolan G., Tavaré S., Dean D., Martin D.H., Bolan G., Tavaré S., Dean D., Bolan G., Tavaré S., Dean D., Tavaré S., Dean D., Dean D. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J. Bacteriol. 2004;186:2457–2465. doi: 10.1128/JB.186.8.2457-2465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman K., Black C.M., Stamm W.E., Jones R.B., Hook E.W., III, Martin D.H., Bolan G., Tavare S., Dean D., Black C.M., Stamm W.E., Jones R.B., Hook E.W., III, Martin D.H., Bolan G., Tavare S., Dean D., Stamm W.E., Jones R.B., Hook E.W., III, Martin D.H., Bolan G., Tavare S., Dean D., Jones R.B., Hook E.W., III, Martin D.H., Bolan G., Tavare S., Dean D., Hook E.W., III, Martin D.H., Bolan G., Tavare S., Dean D., Martin D.H., Bolan G., Tavare S., Dean D., Bolan G., Tavare S., Dean D., Tavare S., Dean D., Dean D. Population-based genetic epidemiologic analysis of Chlamydia trachomatis serotypes and lack of association between ompA polymorphisms and clinical phenotypes. Microbes Infect. 2006;8:604–611. doi: 10.1016/j.micinf.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Morré S.A., Rozendaal L., van Valkengoed I.G., Boeke A.J., van Voorst Vader P.C., Schirm J., de Blok S., van Den Hoek J.A., van Doornum G.J., Meijer C.J., Rozendaal L., van Valkengoed I.G., Boeke A.J., van Voorst Vader P.C., Schirm J., de Blok S., van Den Hoek J.A., van Doornum G.J., Meijer C.J., van Valkengoed I.G., Boeke A.J., van Voorst Vader P.C., Schirm J., de Blok S., van Den Hoek J.A., van Doornum G.J., Meijer C.J., Boeke A.J., van Voorst Vader P.C., Schirm J., de Blok S., van Den Hoek J.A., van Doornum G.J., Meijer C.J., van Voorst Vader P.C., Schirm J., de Blok S., van Den Hoek J.A., van Doornum G.J., Meijer C.J., Schirm J., de Blok S., van Den Hoek J.A., van Doornum G.J., Meijer C.J., de Blok S., van Den Hoek J.A., van Doornum G.J., Meijer C.J., van Den Hoek J.A., van Doornum G.J., Meijer C.J., van Doornum G.J., Meijer C.J., Meijer C.J., et al. Urogenital Chlamydia trachomatis serovars in men and women with a symptomatic or asymptomatic infection: An association with clinical manifestations? J. Clin. Microbiol. 2000;38:2292–2296. doi: 10.1128/jcm.38.6.2292-2296.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Kumar S., Kumar S. Molecular evolution and phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- Ogata H., Renesto P., Audic S., Robert C., Blanc G., Fournier P.E., Parinello H., Claverie J.M., Raoult D., Renesto P., Audic S., Robert C., Blanc G., Fournier P.E., Parinello H., Claverie J.M., Raoult D., Audic S., Robert C., Blanc G., Fournier P.E., Parinello H., Claverie J.M., Raoult D., Robert C., Blanc G., Fournier P.E., Parinello H., Claverie J.M., Raoult D., Blanc G., Fournier P.E., Parinello H., Claverie J.M., Raoult D., Fournier P.E., Parinello H., Claverie J.M., Raoult D., Parinello H., Claverie J.M., Raoult D., Claverie J.M., Raoult D., Raoult D. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 2005;3:e248. doi: 10.1371/journal.pbio.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read T.D., Myers G.S., Brunham R.C., Nelson W.C., Paulsen I.T., Heidelberg J., Holtzapple E., Khouri H., Federova N.B., Carty H.A., Myers G.S., Brunham R.C., Nelson W.C., Paulsen I.T., Heidelberg J., Holtzapple E., Khouri H., Federova N.B., Carty H.A., Brunham R.C., Nelson W.C., Paulsen I.T., Heidelberg J., Holtzapple E., Khouri H., Federova N.B., Carty H.A., Nelson W.C., Paulsen I.T., Heidelberg J., Holtzapple E., Khouri H., Federova N.B., Carty H.A., Paulsen I.T., Heidelberg J., Holtzapple E., Khouri H., Federova N.B., Carty H.A., Heidelberg J., Holtzapple E., Khouri H., Federova N.B., Carty H.A., Holtzapple E., Khouri H., Federova N.B., Carty H.A., Khouri H., Federova N.B., Carty H.A., Federova N.B., Carty H.A., Carty H.A., et al. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): Examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 2003;31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridderhof J.C., Barnes R.C., Barnes R.C. Fusion of inclusions following superinfection of HeLa cells by two serovars of Chlamydia trachomatis. Infect. Immun. 1989;57:3189–3193. doi: 10.1128/iai.57.10.3189-3193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D.L., Hahn B.H., Sharp P.M., Hahn B.H., Sharp P.M., Sharp P.M. Recombination in AIDS viruses. J. Mol. Evol. 1995;40:249–259. doi: 10.1007/BF00163230. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salminen M.O., Carr J.K., Burke D.S., McCutchan F.E., Carr J.K., Burke D.S., McCutchan F.E., Burke D.S., McCutchan F.E., McCutchan F.E. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- Smith J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- Smith J.M., Smith N.H., O’Rourke M., Spratt B.G., Smith N.H., O’Rourke M., Spratt B.G., O’Rourke M., Spratt B.G., Spratt B.G. How clonal are bacteria? Proc. Natl. Acad. Sci. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R.S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Marathe R., Aravind L., Mitchell W., Olinger L., Aravind L., Mitchell W., Olinger L., Mitchell W., Olinger L., Olinger L. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Stothard D.R., Toth G.A., Batteiger B.E., Toth G.A., Batteiger B.E., Batteiger B.E. Polymorphic membrane protein H has evolved in parallel with the three disease-causing groups of Chlamydia trachomatis. Infect. Immun. 2003;71:1200–1208. doi: 10.1128/IAI.71.3.1200-1208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson N.R., Yeats C., Bell K., Holden M.T., Bentley S.D., Livingstone M., Cerdeno-Tarraga A.M., Harris B., Doggett J., Ormond D., Yeats C., Bell K., Holden M.T., Bentley S.D., Livingstone M., Cerdeno-Tarraga A.M., Harris B., Doggett J., Ormond D., Bell K., Holden M.T., Bentley S.D., Livingstone M., Cerdeno-Tarraga A.M., Harris B., Doggett J., Ormond D., Holden M.T., Bentley S.D., Livingstone M., Cerdeno-Tarraga A.M., Harris B., Doggett J., Ormond D., Bentley S.D., Livingstone M., Cerdeno-Tarraga A.M., Harris B., Doggett J., Ormond D., Livingstone M., Cerdeno-Tarraga A.M., Harris B., Doggett J., Ormond D., Cerdeno-Tarraga A.M., Harris B., Doggett J., Ormond D., Harris B., Doggett J., Ormond D., Doggett J., Ormond D., Ormond D., et al. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 2005;15:629–640. doi: 10.1101/gr.3684805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.P., Grayston J.T., Grayston J.T. Serotyping of Chlamydia trachomatis by indirect fluorescent-antibody staining of inclusions in cell culture with monoclonal antibodies. J. Clin. Microbiol. 1991a;29:1295–1298. doi: 10.1128/jcm.29.7.1295-1298.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.P., Grayston J.T., Grayston J.T. Three new serovars of Chlamydia trachomatis: Da, Ia, and L2a. J. Infect. Dis. 1991b;163:403–405. doi: 10.1093/infdis/163.2.403. [DOI] [PubMed] [Google Scholar]
- Workowski K.A., Stevens C.E., Suchland R.J., Holmes K.K., Eschenbach D.A., Pettinger M.B., Stamm W.E., Stevens C.E., Suchland R.J., Holmes K.K., Eschenbach D.A., Pettinger M.B., Stamm W.E., Suchland R.J., Holmes K.K., Eschenbach D.A., Pettinger M.B., Stamm W.E., Holmes K.K., Eschenbach D.A., Pettinger M.B., Stamm W.E., Eschenbach D.A., Pettinger M.B., Stamm W.E., Pettinger M.B., Stamm W.E., Stamm W.E. Clinical manifestations of genital infection due to Chlamydia trachomatis in women: Differences related to serovar. Clin. Infect. Dis. 1994;19:756–760. doi: 10.1093/clinids/19.4.756. [DOI] [PubMed] [Google Scholar]
- World Health Organization, Global prevalence and incidence of selected curable sexually transmitted infections overview and estimates. World Health Organization; Geneva: 2001. [Google Scholar]
- Yang Z., Nielsen R., Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]