Abstract
All Salmonella can cause disease but severe systemic infections are primarily caused by a few lineages. Paratyphi A and Typhi are the deadliest human restricted serovars, responsible for ∼600,000 deaths per annum. We developed a Bayesian changepoint model that uses variation in the degree of nucleotide divergence along two genomes to detect homologous recombination between these strains, and with other lineages of Salmonella enterica. Paratyphi A and Typhi showed an atypical and surprising pattern. For three quarters of their genomes, they appear to be distantly related members of the species S. enterica, both in their gene content and nucleotide divergence. However, the remaining quarter is much more similar in both aspects, with average nucleotide divergence of 0.18% instead of 1.2%. We describe two different scenarios that could have led to this pattern, convergence and divergence, and conclude that the former is more likely based on a variety of criteria. The convergence scenario implies that, although Paratyphi A and Typhi were not especially close relatives within S. enterica, they have gone through a burst of recombination involving more than 100 recombination events. Several of the recombination events transferred novel genes in addition to homologous sequences, resulting in similar gene content in the two lineages. We propose that recombination between Typhi and Paratyphi A has allowed the exchange of gene variants that are important for their adaptation to their common ecological niche, the human host.
Most bacteria undergo frequent homologous recombination, whereby portions of their genomes are replaced by the corresponding sequences from other bacteria (Smith et al. 1993; Suerbaum et al. 1998; Brown et al. 2003; Jolley et al. 2005). Homologous recombination can result in novel combinations of cell surface antigens that facilitate immune escape during epidemic spread within human populations in Neisseria meningitidis (Linz et al. 2000; Zhu et al. 2001), but in general the role of recombination in the evolution of other phenotypic adaptations remains unclear, and its extent has not been quantified at the genomic level. Indeed, it has even been suggested that homologous recombination is a side effect of DNA uptake as a nutrient source, or for the selfish transmission of bacteriophages (Redfield 2001), and need not be directly relevant to host adaptation and pathogenicity.
Here we investigate the extent and origin of recombination that has occurred during the evolution of the two most important human-specific serovars of Salmonella enterica, Typhi and Paratyphi A. Most S. enterica, e.g., the widespread serovars Typhimurium and Enteritidis, colonize the mucosal surfaces of a wide range of mammals and birds. However, some serovars have become specialized, host-specific pathogens that can cause systemic disease, such as typhoid fever in humans (Uzzau et al. 2000). We have taken advantage of the availability of genome sequences of Salmonella Paratyphi A strain ATCC 9150 (McClelland et al. 2004), Typhi strain CT18 (Parkhill et al. 2001), and other serovars, as well as seven housekeeping gene fragments sequenced from diverse S. enterica to investigate the evolution of these two virulent lineages.
Results
Genomic pattern of relatedness between Typhi and Paratyphi A
Genome-wide recombination patterns were investigated using a novel statistical algorithm that infers genetic exchange on the basis of the distribution of nucleotide differences between pairs of strains. If the ancestor of one of the strains imported DNA from a close relative of the other, then that should result in a stretch of DNA with high sequence homology. Alternatively, DNA imported from an unrelated strain might instead exhibit an atypically high level of nucleotide divergence compared with the neighboring region. Our algorithm detects points in the genomic sequences where divergence levels change, which should correspond to the beginning and end points of specific imports, assuming that the neutral levels of polymorphism between the strains are uniform (Hughes and Friedman 2004).
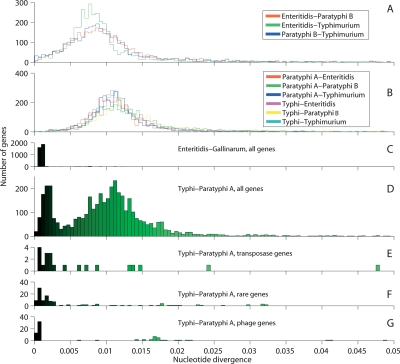
The genomes of S. enterica Typhi, Paratyphi A, Enteritidis, Paratyphi B, Typhimurium, Choleraesuis (Chiu et al. 2005), Infantis, and Hadar were aligned and tested in pairwise fashion by our changepoint model in order to estimate divergence rates for each gene. In all but one of the 64 pairwise comparisons, the divergence levels were characterized by a unimodal distribution with a broad peak at ∼1.0% (Fig. 1A,B; Supplemental Fig. S1). In contrast, the comparison of Typhi versus Paratyphi A yielded a bimodal distribution, with peaks at 0.18% and 1.2% (Fig. 1D), containing ∼23% and 77% of the genes, respectively. The regions constituting these two peaks are subsequently referred to as low- and high-divergence regions. The comparison of Enteritidis and its subserovar Gallinarum yields a unimodal distribution with a peak at 0.1%, similar to the low-divergence regions between Typhi and Paratyphi A (Fig. 1C). Thus, the high-divergence regions of Typhi and Paratyphi A are as distant as random serovars while the low-divergence regions are almost as similar as closely related subserovars.
Figure 1.
Gene-by-gene divergence levels between pairs of genomes of Salmonella enterica. The histograms show the distribution of divergence levels for each gene as estimated by the changepoint model. The intensity of green is proportional to the divergence level up to 2% and is used in Figure 2. Pairwise comparisons, showing respectively unrelated S. enterica genomes excluding Typhi or Paratyphi A (A), the same genomes with Typhi and Paratyphi A (B) and of the closely related Enteritidis and Gallinarum genomes (C). The Typhi versus Paratyphi A comparison showing all genes (D) and homologous transposase genes (E), rare genes (F), and phage genes (G).
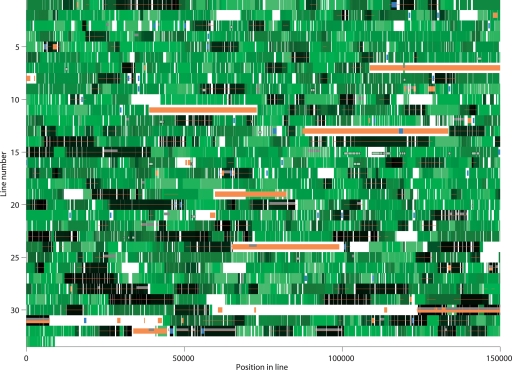
In total there are 124 low-divergence regions in the comparison of Typhi and Paratyphi A, with an average size of 6.4 kb and containing a total of 948 genes (Fig. 2). The high-divergence regions are on average 32 kb in size. After accounting for spatial clustering of genes with a similar function, we find that most classes of genes are randomly distributed between the low- and high-divergence regions (Supplemental Table S1). The only classes of genes that are obviously enriched within the low-divergence regions are shared “rare genes” and genes encoding transposases, as detailed below.
Figure 2.
Divergeome of Typhi in comparison with Paratyphi A. The Typhi genome is shown in 32 lines of 150 kb each and one line of 9037 bp. The starting point is as published in Parkhill et al. (2001). Each gene is color-coded by divergence level in green, as in Figure 1D. Regions where Paratyphi A does not align are shown in white. The low-divergence regions are indicated by a red line. Phage genes are shown in orange, transposase genes in blue, and rare genes in gray. Genes for which Typhi is <0.3% diverged at the nucleotide level to one of the seven test genomes are shown as white boxes with black frames. A full list of genes and their positions is shown in Supplemental Table S4. An equivalent divergeome for Paratyphi A is shown in Supplemental Fig. S2.
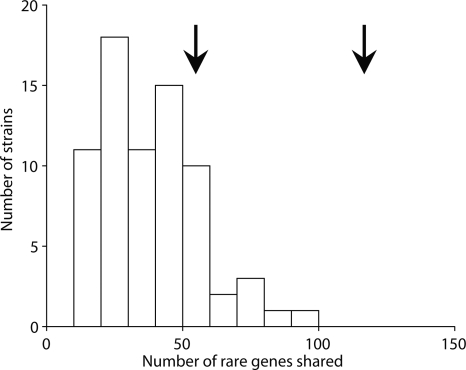
A microarray study (Porwollik et al. 2004) based on the genes found in the Typhi genome revealed that 456 of them are “rare,” defined as being found in <30 of a panel of 78 strains in 27 serovars. We excluded from our analysis 85 of these rare genes which were found in the large SPI7 pathogenicity island, because they are likely to be inherited in a single block (Pickard et al. 2003), resulting in 129 shared rare genes in common. The Paratyphi A genome contains 129 rare genes of Typhi grouped in 57 clusters, which is more than other serovars do (Fig. 3). However, 83 of these shared genes in 29 clusters are in low-divergence regions (Fig. 2). This is significantly more than random expectation (P < 0.005, Fisher’s exact test). Indeed, if the low-divergence regions had the same frequency of rare shared genes as the high-divergence regions, then Paratyphi A would not be atypically close to Typhi in gene content (Fig. 3).
Figure 3.
Histogram of the number of rare genes (as defined in the main text) shared by Typhi CT18 and other strains of S. enterica. The arrow at 120 shows the number of genes shared between Typhi and Paratyphi A while the arrow at 55 shows the projected number of genes if the whole genome had the same frequency of rare genes as the high-divergence regions. Note that we have excluded the large SPI7 region, which is shared between Typhi and a handful of other strains. The data is from Porwollik et al. (2004).
Twenty-one of the 50 transposase genes in Typhi are at syntenic locations in the Paratyphi A genome, and 13 of these (62%) are located in regions of low divergence (Figs. 1E, 2), which is significantly higher than random expectations (P = 0.001, Fisher’s exact test). Transposons jump from one genomic region to another and are also lost with a certain frequency (Beuzon et al. 2004), which reduces the number of transposases found at syntenic positions over evolutionary time. The rate of occurrence of syntenic transposase genes is about five times higher in the low-divergence regions (13 × 0.77/8 × 0.23 = 5.44), which are about five times less diverged than the high-divergence regions (1.2/0.18 = 6.6). This result supports the idea that the overrepresentation of syntenic transposase genes within low-divergence regions is a reflection of loss of syntenic transposons according to a molecular clock analogous to the one that governs sequence divergence.
It has previously been suggested that the independent degradation of certain genes or genes within certain pathways has been important for the convergent phenotypes of Typhi and Paratyphi A (Parkhill et al. 2001; McClelland et al. 2004). Of the 28 genes which are degraded in both strains, six are found in low-divergence regions, which does not deviate significantly from random expectation. Of these six, three are transposase remnants, one (dmsA) carries independent inactivating mutations in the two genomes, and two (sugR and sopA) have the same initial inactivating mutation in Typhi and Paratyphi A, implying common ancestry. Thus, on its own, the presence of the low-divergence regions does not account for the similarities observed in genome degradation.
There are three prophages in the Paratyphi A genome and seven in Typhi. One of the three found in Paratyphi A, SPA-2-SopE, is highly similar to the SopE phage in Typhi, in fact representing the region of the genome with the lowest level of divergence (Fig. 1G; lines 30–31 on Fig. 2). The other two Paratyphi A phages show only incomplete, limited homology with phages in Typhi.
In summary we have found that, although according to several different criteria the Typhi and Paratyphi A genomes are more similar to each other than to the other members of S. enterica that have been sequenced, this similarity is almost entirely due to a very high degree of homology of a quarter of the genome. The other three quarters are about as different as randomly selected strains in terms of gene content and nucleotide divergence. How did the two genomes come to have regions with such distinct evolutionary histories?
Comparison of pattern of relatedness with expectations under two evolutionary scenarios
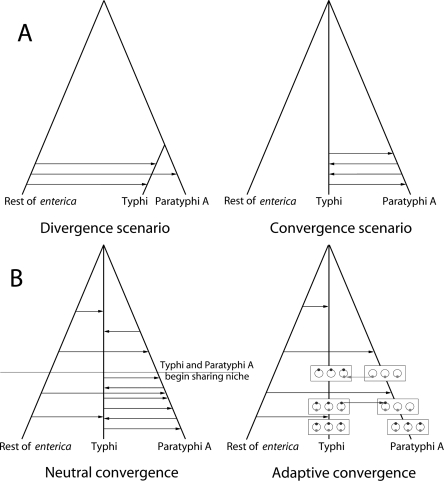
The striking bimodality of the distribution in the Typhi versus Paratyphi A comparison could potentially be explained by two quite different scenarios (Fig. 4A). Under the divergence scenario, Typhi and Paratyphi A share a recent common ancestor. The relatively recent separation would then be reflected by the low-divergence regions (0.18% nucleotide distance), which would correspond to segments of the genome that have not recombined. The majority of the genome would, however, have been imported from other S. enterica into either Typhi or Paratyphi A, resulting in the ∼1% divergence value that is typically observed between unrelated serovars (Fig. 1A,B).
Figure 4.
Scenarios for the divergence of Typhi and Paratyphi A. The trees indicate the clonal history and the arrows indicate genetic exchange events. (A) Illustration of the two scenarios that can explain the bimodal distribution of divergence levels (Fig. 1D). (B) Illustration of the two convergence factors. Under neutral convergence, the two lineages converge due to an elevated rate of exchange between them. Under adaptive convergence, exchanges between the two lineages are preferentially fixed because they contain alleles (gray dots), which confer a selective advantage in their common niche.
Under the convergence scenario, the ancestors of Typhi and Paratyphi A were as unrelated as are most other pairs of serovars of S. enterica, but 23% of their genome in total have been recently imported from one lineage to the other. The remaining 77% of the genome would then either reflect the original composition of the lineages prior to this recombination or have been imported from other serovars. The convergence scenario requires much less recombination in total than under divergence and, for this reason, is intuitively more appealing. Frequent recombination involving DNA donors outside of S. enterica subspecies enterica is unlikely according to both scenarios because the resulting nucleotide differences would be >1.2%.
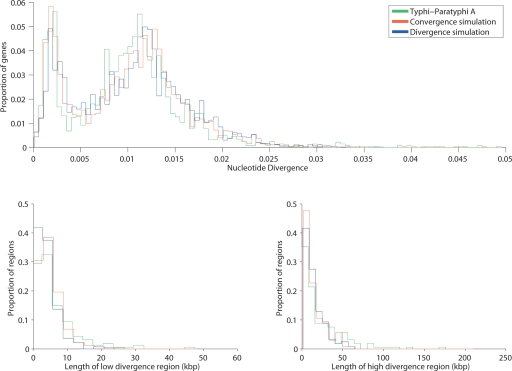
We performed simulations in order to see whether the observed spatial distribution of low- and high-divergence regions was consistent with what is expected under the two scenarios. Although the two scenarios can each give quite distinct signatures in some circumstances (e.g., shortly after convergence), we were able to obtain a good fit to the observed distribution of the lengths of the low- and high-divergence regions under both scenarios (Fig. 5) by adjusting average tract length and other parameters (see Methods).
Figure 5.
Properties of low- and high-divergence regions in Typhi vs. Paratyphi A, compared with genomes simulated according to convergence and divergence scenarios.
These simulations showed that, because recombination events sometimes overlap, the actual number of imports may have been much higher than the observed number of boundaries. The simulations in Figure 5 involved 870 imports in the convergence scenario and 3979 imports under divergence. The mean tract length in both cases was 1700 base pairs. In both simulations, a constant rate of 0.18% nucleotide differences was used for the low divergence regions. This gives a better fit than using a uniform distribution between 0% and 0.3% (data not shown). This observation is expected under the divergence scenario, but is also consistent with convergence if recombination occurred within a short time span.
Although the spatial distribution of low and high divergence is consistent with both scenarios, the pattern that we observe between Paratyphi A and Typhi is qualitatively and quantitively different from the patterns observed in the recent divergence between Enteritidis and Gallinarum (Fig. 1, cf. C and D). The Enteritidis and Gallinarum genomes differ by approximately half as many mutations per kilobase as observed in the low-divergence region of Typhi and Paratyphi A, but the fraction of the genome in high-divergence regions is 20-fold lower. The divergence scenario would, therefore, require a 10-fold higher rate of recombinational divergence between Typhi and Paratyphi A than observed between Enteritidis and Gallinarum. Moreover, most of the high-divergence regions between Enteritidis and Gallinarum are contiguous (not shown), apparently reflecting one large event, as opposed to more than a hundred high-divergence regions in the Typhi and Paratyphi A comparison. The pattern of import that would be required under the divergence scenario is, therefore, quantitatively and qualitatively different from the one that took place in the divergence of Enteritidis and Gallinarum, making this scenario less likely.
Possible markers of recombination
Some parts of the genome may be more susceptible to homologous recombination than others. We would expect these regions to be overrepresented in the parts of the genome that recombined most recently, i.e., the low-divergence region under the convergence hypothesis and the high-divergence region under the divergence hypothesis. For example, since prophages encode their own means of transmission from one bacteria to another, they might be expected to recombine more frequently than the rest of the genome. As described above, one of the three prophages of Paratyphi A is that element of the genome which is most closely related to Typhi, while the other two are unrelated.
χ sequences prevent the RecBCD enzyme from digesting DNA within the cell and thus may facilitate homologous recombination of the stretches that contain them (Miesel and Roth 1994; Handa et al. 1997). There were 846 χ sequences in the Typhi genome, of which 140 were in low-divergence regions of the alignment, which is not significantly high or low (P = 0.2) considering the slightly lower GC content in low-divergence regions (51% compared with 52% in high-divergence regions). Moreover, χ sequences are not over- or underrepresented in the 200 base pairs on either of side of the boundaries between low- or high-divergence regions, as would be expected if they played a substantial role in facilitating the exchange of DNA.
Evidence from MLST data
For many organisms, convergence and divergence could be distinguished by comparisons of related isolates on the basis of sequences from several housekeeping gene fragments (multilocus sequence typing [MLST]; Maiden et al. 1998). However, according to MLST, very little genetic diversity was found among 26 strains of Typhi due to a recent bottleneck (Kidgell et al. 2002). The only three changes that were observed were single nucleotide changes probably due to mutation. Data for 28 Paratyphi A strains at the S. enterica MLST database (http://web.mpiib-berlin.mpg.de/mlst/) indicate that Paratyphi A is similarly uniform. Additionally, there were no close relatives of either Typhi or Paratyphi A among 597 other Salmonella in the MLST database. No non-Typhi strain possessed a single Typhi allele and no non-Paratyphi A strain possessed more than two Paratyphi A alleles. Thus current patterns of S. enterica diversity as assayed by MLST provide no clue as to the nature of the substantial amount of recombination that took place in the history of these lineages.
Genetic imports from other members of S. enterica
The two scenarios make contradictory predictions about genetic relationships expected for imported fragments. Under the convergence hypothesis, recent recombination has been between Typhi and Paratyphi A and should not affect relationships with other strains. Under the divergence hypothesis, three quarters of the genome would have been imported from other S. enterica, much of it quite recently, and it should therefore be possible to identify relatives of the strains from which Typhi or Paratyphi A imported DNA.
We used genomes from seven serovars (Enteritidis, Typhimurium, Paratyphi B, Dublin, Choleraesuis, Hadar, and Infantis) to search for putative imports required by the divergence scenario. Only very few Paratyphi A or Typhi genes can be attributed to import from one of these seven serovars. Except for 106 kb (3% of the genome), all other coding sequences differed by >0.3% estimated divergence from their orthologs in all seven genomes (Supplemental Table S2). Many of these homologous regions probably do not represent true imports from the seven lineages. For example, one homologous stretch included a cluster of 12 ribosomal RNA genes that showed low divergence in all pairwise comparisons, with the lowest pairwise difference being between Paratyphi A and Typhi. Other putative imports were too short to be convincing (<1 kb) and probably represent false positives. Only two to four regions may represent true imports, totaling 10–15 kb (Supplemental Table S2).
Thus, <0.5% of the imports needed under the divergence hypothesis can be attributed to close relatives of Enteritidis, Typhimurium, Paratyphi B, Dublin, Choleraesuis, Infantis, or Hadar. Under the divergence hypothesis, we should have detected a much higher number of imports from one of the seven serovars. These serovars make up a substantial proportion of the S. enterica gene pool, at least according to current snapshots provided by serotyping and MLST of diverse strain collections. The seven serovars represent >50% of Salmonella isolates from chickens, pigs, cattle, and humans in the Netherlands (van Duijkeren et al. 2002) and the USA. (Centers for Disease Control 2005). Furthermore, based on the seven housekeeping gene fragments used for MLST, 68% of the strains with one of the seven serovars, and 29% of all S. enterica strains in the MLST database at the time of analysis, diverged by <0.3% from the corresponding alleles in the genomes that have been sequenced for each of these four serovars.
The frequency of specific lineages and serovars can change over time and our knowledge about Salmonella is doubtless biased toward humans and agricultural reservoirs, so that the currently common lineages in our databases might constitute only part of the historical gene pool. This argument seems unlikely to explain our inability to identify a source of imports: The seven test genomes exhibit significantly more genetic exchange with each other than with either Typhi or Paratyphi A, with a higher proportion of the pairwise comparisons between the seven test genomes showing divergence levels <0.3% (Fig. 1, cf. A and B; Supplemental Fig. S1, cf. A and B). Secondly, for the six MLST gene fragments that are located in high-divergence regions, the MLST database did not contain any strains with alleles more closely related to those of Typhi or Paratyphi A than those of the seven test genomes (Supplemental Table S3). Thus, the seven test genomes are as good, or better, than any other candidate in the MLST database as a source of genes for importation in the divergence scenario. We, therefore, reject the divergence scenario and infer that Typhi and Paratyphi A have converged by homologous recombination.
Discussion
Summary of findings
We have described a surprising pattern of genomic relatedness between Typhi and Paratyphi A. Three quarters of their genomes show a level of genetic similarity that is typical of distantly related members of the species S. enterica, both in gene content and nucleotide divergence. The remaining quarter, however, is much more similar in both aspects, with average nucleotide divergence of 0.18% instead of 1.2%.
There are two quite different scenarios that could explain this pattern, recent convergence or recent divergence. Both explanations require an atypical and unclock-like pattern of recombination, when compared with other S. enterica. We rejected the divergence hypothesis. The divergence hypothesis would require several-fold more recombination events with other members of S. enterica than convergence. This rate of recombination would also have needed to be much higher for Typhi and Paratyphi A than for Enteritidis and Gallinarum. Most importantly, we were unable to identify a plausible origin of the imported genes amongst seven other sequenced genomes of S. enterica, although these are as similar to Typhi and Paratyphi A as any of almost 600 strains that have been MLST-typed.
We infer that convergence has taken place, which implies an extremely elevated rate of recombination between Typhi and Paratyphi A. That spate of recombination probably occurred over a short period that preceded the most recent population bottlenecks in the history of Typhi and Paratyphi A. Only a few genes diverge between these genomes by <0.1%, with a clear mode at 0.18% that presumably reflects the accumulation of mutations subsequent to recombination. In comparison, the bottleneck within Typhi is associated with average synonymous sequence variation of ∼0.05% (Kidgell et al. 2002).
Role of selection
There is an attractive evolutionary argument for the convergence scenario because the two lineages have adapted to the same spatial and ecological niche: obligate, invasive infection of the human host. Typhi and Paratyphi A invade the same tissues and there are even rare reported cases of coinfection (Joshi et al. 2002), which may increase the opportunity for genetic exchange (neutral convergence, Fig. 4B). Furthermore, in adapting to the specific lifestyle of pathogenic transmission, the two bacteria will have had to learn the same tricks, possibly from one another, which could promote the fixation of specific imports (adaptive convergence, Fig. 4B).
However, despite its evolutionary logic, it is difficult to find unambiguous evidence for the importance of selection. Each of the 124 low-divergence regions contains several genes. Adaptive convergence would require only a fraction of these to be positively selected, which may explain why we were unable to find any overrepresentation of particular categories of genes within the low-divergence regions (Supplemental Table S1).
One particularly interesting feature of the low divergence regions is that they contain an overrepresentation of rare genes shared between Typhi and Paratyphi A, compared both with the high-divergence regions and with the number of rare genes shared between Typhi and other strains (Figs. 2, 3). The 83 shared rare genes found in homologous low-divergence regions thus represent particularly good candidates for adaptive convergence. Unfortunately, the existence of these shared genes does not indicate the directionality of exchange or prove the role of selection.
Means of convergence
Due to the uniformity of the divergence rate within the low-divergence regions, we argue that convergence between Typhi and Paratyphi A happened in a limited time span and then stopped. But how could recombination between a pair of lineages occur in such a rapid burst? One speculative explanation is that recombination was mediated by epidemic infection of both lineages by a bacteriophage capable of generalized transduction. Generalized transduction occurs by the incorporation of bacterial DNA instead of phage DNA into the viral capsid and is often a property of lytic phages (Schicklmaier and Schmieger 1995). None of the prophages found in either Typhi or Paratyphi A are obvious candidates for such a role and the only shared prophage, SopEΦ, is poor at transduction because it efficiently excludes host DNA from the phage head.
Our inability to detect a suitable candidate phage within the genome of Typhi or Paratyphi A is not crucial. Firstly, if the putative phage was lyric it would not have integrated into the host cell genome. Secondly, the fact that recombination occurred over a short time span suggests that the phage was lost in a bottleneck subsequent to its epidemic spread. Thirdly, one or both lineages may have evolved resistance to the phage. Indeed, the most closely related genes within the two genomes encode SopEΦ. SopEΦ has genes of unknown function at syntenic locations where the well characterized phage exclusion genes tin and old are found in P2 (Pelludat et al. 2003) that might also cause phage exclusion. Thus, integration of SopEΦ may have marked the end of the epidemic spread of lytic phages and associated transduction.
This scenario implies that a transient mechanism or transient ecological conditions, such as high coinfection rates, can facilitate highly nonrandom patterns of recombination. This recombination can in turn lead to a saltation in which a hybrid strain or strains are rapidly assembled from the constituent parts of two or more lineages. Hybridization has been shown to facilitate the evolution of novel and extreme phenotypes in animal (Schwarz et al. 2005), plant (Rieseberg et al. 2003), and virus (Shaw et al. 2002) systems, and a recent study has suggested that an emergent virulent clone of the bacteria Vibrio vulnificus was a hybrid between two major gene pools (Bisharat et al. 2005).
Conclusions
How would it be possible to find definitive evidence for convergence and also for the potential role of a bacteriophage or natural selection? Any singular evolutionary event presents a challenge for scientific analysis, particularly if it is no longer ongoing, but convincing proof might nevertheless be found. Firstly, if other S. enterica could be identified that were distant relatives of Typhi and Paratyphi A, perhaps using more extensive sequence data sets, then this would prove convergence. Secondly, similar events might be identified in other bacterial lineages that have left more distinctive signatures. Thirdly there may still be untapped information in the extant genomes of Typhi and Paratyphi A on current patterns of evolution that will inform us about historical processes we are seeking to understand.
Whatever mechanism led to the convergence of Typhi and Paratyphi A, our results highlight the potential importance of homologous recombination as an evolutionary force, in addition to the more widely recognized mechanisms of lateral gene transfer and gene degradation. They also demonstrate that genomic patterns of recombination can be highly nonrandom, even within a single species of bacteria, which has important consequences for our understanding of bacterial evolution and the evolution of pathogenicity.
Methods
Genomes
In addition to Paratyphi A ATCC 9150 (McClelland et al. 2004) and Typhi CT18 (Parkhill et al. 2001), the strains Choleraesuis (Chiu et al. 2005), Paratyphi B SPB7 (http://genome.wustl.edu/), Dublin (http://www.salmonella.org/genomics/http://www.salmonella.org/genomics/), Enteritidis PT4, Typhimurium DT104, Hadar, and Infantis (all four from http://www.sanger.ac.uk/Projects/Salmonella) were chosen as unrelated genomes within S. enterica. The strain of Gallinarum 287/91 (http://www.sanger.ac.uk/Projects/Salmonellahttp://www.sanger.ac.uk/Projects/Salmonella) was chosen as a relative of Enteritidis PT4.
Alignments
All alignments were made using MAUVE (Darling et al. 2004). Figure 1A,B is based on a partial multiple alignment of Typhi, Paratyphi A, Enteritidis, Typhimurium, and Paratyphi B, whereas Figures 1D–G, 2, 5, and Supplemental Figure S2 are based on a pairwise alignment of the Paratyphi A and Typhi genomes. Figure 1C is based on a pairwise alignment of Enteritidis and Gallinarum. Putative imports in Supplemental Table S2 between Dublin and Paratyphi A or Typhi were searched for using a three-way alignment of these sequences, as for the imports from Hadar, Infantis, and Choleraesuis.
The prior model
In a Bayesian framework, the prior specifies our initial expectation of the structure of relationships between two aligned genomic sequences, before observation. An alignment consists of locally collinear blocks of homologous sequences. Here we consider only one block for clarity, but the same prior model holds for each block and our algorithm runs on all blocks simultaneously. Our prior model assumes that the alignment is made of a certain number k of segments, within which the level of nucleotide divergence between the two genomes is constant. Similar assumptions for a block-like structure of the nucleotide divergence level are made and discussed in Suchard et al. (2003) or Husmeier and McGuire (2003). The prior on k is a binomial distribution whose mean is estimated from the data, with a uniform hyperprior on this mean. The location of the boundaries between segments is uniform given k. The levels of divergence within each segment are assumed to be independent, identically distributed draws from the same Beta prior with hyperparameters estimated from the data. This is a convenient choice given that the observed number of nucleotide differences within each segment is binomially distributed and that the Beta distribution is the conjugate of the Binomial distribution. Moreover, the Beta distribution is a flexible distribution which can take many forms depending on its parameters.
Bayesian inference
Here our general approach is to use a Monte Carlo Markov Chain to generate a succession of values for each parameter which converges on the posterior, i.e., our beliefs about the parameters after observation of the data. Each parameter (or block of parameters) is sequentially updated by a new value drawn from a proposal distribution depending on the remaining parameters and the data. Under the Metropolis-Hastings scheme, proposed new parameters are accepted with a probability that depends on the likelihood of the new parameters relative to the existing ones. The difficulty here is that the number of parameters depends on the number of segments k which is unknown. We use reversible jump MCMC (Green 1995) in which a type of chain jumps between different values of k, while preserving most of the information held in the remaining parameters. Our algorithm uses two transdimensional jump proposals. The first is to increase the value of k by splitting a randomly chosen segment at a randomly chosen position and giving the two proposed segments a homology drawn from their posterior distribution conditional on the other parameters. The second decreases the value of k by merging two consecutive segments and giving the new merged segment a level of homology drawn from its conditional posterior distribution. In addition, we use two non-transdimensional moves. The first proposes moving the boundary to another location. The second updates the level of homology of a chosen segment. Finally, the hyperparameters of the prior model are updated periodically given the rest of the parameters. Each chain was run for at least 10,000,000 iterations and repeated to check for convergence. The distance attributed to a gene is the mean value of the distance for the segments it belongs to, weighted by their length. The regions of low divergence (red lines in Fig. 2) are defined as contiguous sets of genes for which the distance between Typhi and Paratyphi A is <0.5% and the flanking genes have divergence <0.3%.
Simulation of convergence and divergence scenarios
We simulate the differences between two genomes equal in size to Typhi (Fig. 5). Under divergence, the backbone of the genome has a constant nucleotide divergence of 0.18%, which is assumed to have arisen by point mutation. Imports have higher average divergence levels (normally distributed with mean 1.2% and standard deviation 0.7%), which represents the fact that recombination happens with other S. enterica strains which may be more or less related to the Typhi and Paratyphi A genomes. Imports, whose size is geometrically distributed with mean 1700 bp, are added at random locations on the genome until the backbone represents 23% of the genome. Under convergence, the backbone of the genome is assumed to consist of chunks of variable divergence (mean 1.2% and standard deviation 0.7%), which here is assumed to reflect extensive recombination since the genomes diverged (and is broadly consistent with the pairwise differences observed between other strains). Imports from one lineage to the other, again with mean size 1700 bp, are added at random locations until the background represents 77% of the genome. For both models, each of the parameters was chosen to achieve a good fit with the observed distribution (Fig. 5).
Functional analysis
The annotation of Typhi (Parkhill et al. 2001) was used to identify genes of different functional types in Figure 2 and Supplemental Table S1. Supplemental Figure S2 is based on the Paratyphi A annotation (McClelland et al. 2004). Functional analysis of the Typhi–Paratyphi A alignment was performed using Artemis (Rutherford et al. 2000) and the Artemis Comparison Tool (Carver et al. 2005), both available from http://www.sanger.ac.uk/Software/Artemis/.
Acknowledgments
We thank Rory Bowden, Angus Buckling, Peter Donnelly, Ed Feil, Ichizo Kobayashi, Myrone Levine, Gil McVean, Simon Myers, Phillipe Roumagnac, Brian Spratt, John Wain, and two anonymous reviewers for providing helpful comments or suggestions. This work was funded by the Wellcome Trust.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5512906
References
- Beuzon C.R., Chessa D., Casadesus J., Chessa D., Casadesus J., Casadesus J. IS200: An old and still bacterial transposon. Int. Microbiol. 2004;7:3–12. [PubMed] [Google Scholar]
- Bisharat N., Cohen D.I., Harding R.M., Falush D., Crook D.W., Peto T., Maiden M.C., Cohen D.I., Harding R.M., Falush D., Crook D.W., Peto T., Maiden M.C., Harding R.M., Falush D., Crook D.W., Peto T., Maiden M.C., Falush D., Crook D.W., Peto T., Maiden M.C., Crook D.W., Peto T., Maiden M.C., Peto T., Maiden M.C., Maiden M.C. Hybrid Vibrio vulnificus. Emerg. Infect. Dis. 2005;11:30–35. doi: 10.3201/eid1101.040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.W., Mammel M.K., LeClerc J.E., Cebula T.A., Mammel M.K., LeClerc J.E., Cebula T.A., LeClerc J.E., Cebula T.A., Cebula T.A. Limited boundaries for extensive horizontal gene transfer among Salmonella pathogens. Proc. Natl. Acad. Sci. 2003;100:15676–15681. doi: 10.1073/pnas.2634406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T.J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rajandream M.A., Barrell B.G., Parkhill J., Barrell B.G., Parkhill J., Parkhill J. ACT: The Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control, Salmonella surveillance: Annual summary, 2004. US Department of Health and Human Services; Atlanta, Georgia: 2005. [Google Scholar]
- Chiu C.H., Tang P., Chu C.S., Hu S.N., Bao Q.Y., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Tang P., Chu C.S., Hu S.N., Bao Q.Y., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Chu C.S., Hu S.N., Bao Q.Y., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Hu S.N., Bao Q.Y., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Bao Q.Y., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Chou Y.Y., Wang H.S., Lee Y.S., Wang H.S., Lee Y.S., Lee Y.S. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 2005;33:1690–1698. doi: 10.1093/nar/gki297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A.C.E., Mau B., Blattner F.R., Perna N.T., Mau B., Blattner F.R., Perna N.T., Blattner F.R., Perna N.T., Perna N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P.J. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika. 1995;82:711–732. [Google Scholar]
- Handa N., Ohashi S., Kusano K., Kobayashi I., Ohashi S., Kusano K., Kobayashi I., Kusano K., Kobayashi I., Kobayashi I. Chi*, a chi-related 11-mer sequence partially active in an E coli recC1004 strain. Genes Cells. 1997;2:525–536. doi: 10.1046/j.1365-2443.1997.1410339.x. [DOI] [PubMed] [Google Scholar]
- Hughes A.L., Friedman R., Friedman R. Patterns of sequence divergence in 5′ intergenic spacers and linked coding regions in 10 species of pathogenic bacteria reveal distinct recombinational histories. Genetics. 2004;168:1795–1803. doi: 10.1534/genetics.104.032979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husmeier D., McGuire G., McGuire G. Detecting recombination in 4-taxa DNA sequence alignments with Bayesian hidden Markov models and Markov chain Monte Carlo. Mol. Biol. Evol. 2003;20:315–337. doi: 10.1093/molbev/msg039. [DOI] [PubMed] [Google Scholar]
- Jolley K.A., Wilson D.J., Kriz P., Mcvean G., Maiden M.C.J., Wilson D.J., Kriz P., Mcvean G., Maiden M.C.J., Kriz P., Mcvean G., Maiden M.C.J., Mcvean G., Maiden M.C.J., Maiden M.C.J. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol. Biol. Evol. 2005;22:562–569. doi: 10.1093/molbev/msi041. [DOI] [PubMed] [Google Scholar]
- Joshi S., Wattal C., Sharma A., Prasad K.J., Wattal C., Sharma A., Prasad K.J., Sharma A., Prasad K.J., Prasad K.J. Mixed Salmonella infection—A case report. Indian J. Med. Microbiol. 2002;20:113–114. [PubMed] [Google Scholar]
- Kidgell C., Reichard U., Wain J., Linz B., Torpdahl M., Dougan G., Achtman M., Reichard U., Wain J., Linz B., Torpdahl M., Dougan G., Achtman M., Wain J., Linz B., Torpdahl M., Dougan G., Achtman M., Linz B., Torpdahl M., Dougan G., Achtman M., Torpdahl M., Dougan G., Achtman M., Dougan G., Achtman M., Achtman M. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2002;2:39–45. doi: 10.1016/s1567-1348(02)00089-8. [DOI] [PubMed] [Google Scholar]
- Linz B., Schenker M., Zhu P., Achtman M., Schenker M., Zhu P., Achtman M., Zhu P., Achtman M., Achtman M. Frequent interspecific genetic exchange between commensal Neisseriae and Neisseria meningitidis. Mol. Microbiol. 2000;36:1049–1058. doi: 10.1046/j.1365-2958.2000.01932.x. [DOI] [PubMed] [Google Scholar]
- Maiden M.C., Bygraves J.A., Feil E., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Bygraves J.A., Feil E., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Feil E., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Morelli G., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Russell J.E., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Urwin R., Zhang Q., Zhou J., Zurth K., Caugant D.A., Zhang Q., Zhou J., Zurth K., Caugant D.A., Zhou J., Zurth K., Caugant D.A., Zurth K., Caugant D.A., Caugant D.A., et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Sanderson K.E., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Sanderson K.E., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Meyer R., Bieri T., Ozersky P., McLellan M., Bieri T., Ozersky P., McLellan M., Ozersky P., McLellan M., McLellan M., et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- Miesel L., Roth J.R., Roth J.R. Salmonella recD mutations increase recombination in a short sequence transduction assay. J. Bacteriol. 1994;176:4092–4103. doi: 10.1128/jb.176.13.4092-4103.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J., Dougan G., James K.D., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T.G., Dougan G., James K.D., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T.G., James K.D., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T.G., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T.G., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T.G., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T.G., Churcher C., Mungall K.L., Bentley S.D., Holden M.T.G., Mungall K.L., Bentley S.D., Holden M.T.G., Bentley S.D., Holden M.T.G., Holden M.T.G., et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- Pelludat C., Mirold S., Hardt W.D., Mirold S., Hardt W.D., Hardt W.D. The SopE Phi phage integrates into the ssrA gene of Salmonella enterica serovar typhimurium A36 and is closely related to the Fels-2 prophage. J. Bacteriol. 2003;185:5182–5191. doi: 10.1128/JB.185.17.5182-5191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard D., Wain J., Baker S., Line A., Chohan S., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Wain J., Baker S., Line A., Chohan S., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Baker S., Line A., Chohan S., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Line A., Chohan S., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Chohan S., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Gaora P.O., Chabalgoity J.A., Thanky N., Chabalgoity J.A., Thanky N., Thanky N., et al. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 2003;185:5055–5065. doi: 10.1128/JB.185.17.5055-5065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porwollik S., Boyd E.F., Choy C., Cheng P., Florea L., Proctor E., McClelland M., Boyd E.F., Choy C., Cheng P., Florea L., Proctor E., McClelland M., Choy C., Cheng P., Florea L., Proctor E., McClelland M., Cheng P., Florea L., Proctor E., McClelland M., Florea L., Proctor E., McClelland M., Proctor E., McClelland M., McClelland M. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J. Bacteriol. 2004;186:5883–5898. doi: 10.1128/JB.186.17.5883-5898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield R.J. Do bacteria have sex? Nat. Rev. Genet. 2001;2:634–639. doi: 10.1038/35084593. [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H., Raymond O., Rosenthal D.M., Lai Z., Livingstone K., Nakazato T., Durphy J.L., Schwarzbach A.E., Donovan L.A., Lexer C., Raymond O., Rosenthal D.M., Lai Z., Livingstone K., Nakazato T., Durphy J.L., Schwarzbach A.E., Donovan L.A., Lexer C., Rosenthal D.M., Lai Z., Livingstone K., Nakazato T., Durphy J.L., Schwarzbach A.E., Donovan L.A., Lexer C., Lai Z., Livingstone K., Nakazato T., Durphy J.L., Schwarzbach A.E., Donovan L.A., Lexer C., Livingstone K., Nakazato T., Durphy J.L., Schwarzbach A.E., Donovan L.A., Lexer C., Nakazato T., Durphy J.L., Schwarzbach A.E., Donovan L.A., Lexer C., Durphy J.L., Schwarzbach A.E., Donovan L.A., Lexer C., Schwarzbach A.E., Donovan L.A., Lexer C., Donovan L.A., Lexer C., Lexer C. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B., Horsnell T., Rice P., Rajandream M.A., Barrell B., Rice P., Rajandream M.A., Barrell B., Rajandream M.A., Barrell B., Barrell B. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Schicklmaier P., Schmieger H., Schmieger H. Frequency of generalized transducing phages in natural isolates of the Salmonella typhimurium complex. Appl. Environ. Microbiol. 1995;61:1637–1640. doi: 10.1128/aem.61.4.1637-1640.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D., Matta B.M., Shakir-Botteri N.L., McPheron B.A., Matta B.M., Shakir-Botteri N.L., McPheron B.A., Shakir-Botteri N.L., McPheron B.A., McPheron B.A. Host shift to an invasive plant triggers rapid animal hybrid speciation. Nature. 2005;436:546–549. doi: 10.1038/nature03800. [DOI] [PubMed] [Google Scholar]
- Shaw M., Cooper L., Xu X., Thompson W., Krauss S., Guan Y., Zhou N., Klimov A., Cox N., Webster R., Cooper L., Xu X., Thompson W., Krauss S., Guan Y., Zhou N., Klimov A., Cox N., Webster R., Xu X., Thompson W., Krauss S., Guan Y., Zhou N., Klimov A., Cox N., Webster R., Thompson W., Krauss S., Guan Y., Zhou N., Klimov A., Cox N., Webster R., Krauss S., Guan Y., Zhou N., Klimov A., Cox N., Webster R., Guan Y., Zhou N., Klimov A., Cox N., Webster R., Zhou N., Klimov A., Cox N., Webster R., Klimov A., Cox N., Webster R., Cox N., Webster R., Webster R., et al. Molecular changes associated with the transmission of avian influenza A H5N1 and H9N2 viruses to humans. J. Med. Virol. 2002;66:107–114. doi: 10.1002/jmv.2118. [DOI] [PubMed] [Google Scholar]
- Smith J.M., Smith N.H., O’Rourke M., Spratt B.G., Smith N.H., O’Rourke M., Spratt B.G., O’Rourke M., Spratt B.G., Spratt B.G. How clonal are bacteria? Proc. Natl. Acad. Sci. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M.A., Weiss R.E., Dorman K.S., Sinsheimer J.S., Weiss R.E., Dorman K.S., Sinsheimer J.S., Dorman K.S., Sinsheimer J.S., Sinsheimer J.S. Inferring spatial phylogenetic variation along nucleotide sequences: A multiple changepoint model. J. Am. Stat. Assoc. 2003;98:427–437. [Google Scholar]
- Suerbaum S., Smith J.M., Bapumia K., Morelli G., Smith N.H., Kunstmann E., Dyrek I., Achtman M., Smith J.M., Bapumia K., Morelli G., Smith N.H., Kunstmann E., Dyrek I., Achtman M., Bapumia K., Morelli G., Smith N.H., Kunstmann E., Dyrek I., Achtman M., Morelli G., Smith N.H., Kunstmann E., Dyrek I., Achtman M., Smith N.H., Kunstmann E., Dyrek I., Achtman M., Kunstmann E., Dyrek I., Achtman M., Dyrek I., Achtman M., Achtman M. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzau S., Brown D.J., Wallis T., Rubino S., Leori G., Bernard S., Casadesus J., Platt D.J., Olsen J.E., Brown D.J., Wallis T., Rubino S., Leori G., Bernard S., Casadesus J., Platt D.J., Olsen J.E., Wallis T., Rubino S., Leori G., Bernard S., Casadesus J., Platt D.J., Olsen J.E., Rubino S., Leori G., Bernard S., Casadesus J., Platt D.J., Olsen J.E., Leori G., Bernard S., Casadesus J., Platt D.J., Olsen J.E., Bernard S., Casadesus J., Platt D.J., Olsen J.E., Casadesus J., Platt D.J., Olsen J.E., Platt D.J., Olsen J.E., Olsen J.E. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 2000;125:229–255. doi: 10.1017/s0950268899004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijkeren E., Wannet W.J.B., Houwers D.J., van Pelt W., Wannet W.J.B., Houwers D.J., van Pelt W., Houwers D.J., van Pelt W., van Pelt W. Serotype and phage type distribution of Salmonella strains isolated from humans, cattle, pigs, and chickens in the Netherlands from 1984 to 2001. J. Clin. Microbiol. 2002;40:3980–3985. doi: 10.1128/JCM.40.11.3980-3985.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., van der Ende A., Falush D., Brieske N., Morelli G., Linz B., Popovic T., Schuurman I.G., Adegbola R.A., Zurth K., van der Ende A., Falush D., Brieske N., Morelli G., Linz B., Popovic T., Schuurman I.G., Adegbola R.A., Zurth K., Falush D., Brieske N., Morelli G., Linz B., Popovic T., Schuurman I.G., Adegbola R.A., Zurth K., Brieske N., Morelli G., Linz B., Popovic T., Schuurman I.G., Adegbola R.A., Zurth K., Morelli G., Linz B., Popovic T., Schuurman I.G., Adegbola R.A., Zurth K., Linz B., Popovic T., Schuurman I.G., Adegbola R.A., Zurth K., Popovic T., Schuurman I.G., Adegbola R.A., Zurth K., Schuurman I.G., Adegbola R.A., Zurth K., Adegbola R.A., Zurth K., Zurth K., et al. Fit genotypes and escape variants of subgroup III Neisseria meningitidis during three pandemics of epidemic meningitis. Proc. Natl. Acad. Sci. 2001;98:5234–5239. doi: 10.1073/pnas.061386098. [DOI] [PMC free article] [PubMed] [Google Scholar]