Abstract
Comparative genomic studies have been useful in identifying transcriptional regulatory elements in higher eukaryotic genomes, but many important regulatory elements cannot be detected by such analyses due to evolutionary variations and alignment tool limitations. Therefore, in this study we exploit the highly conserved nature of epigenetic modifications to identify potential transcriptional enhancers. By using a high-resolution genome-wide mapping technique, which combines the chromatin immunoprecipitation and serial analysis of gene expression assays, we have recently determined the distribution of lysine 9/14-diacetylated histone H3 in human T cells. We showed the existence of 46,813 regions with clusters of histone acetylation, termed histone acetylation islands, some of which correspond to known transcriptional regulatory elements. In the present study, we find that 4679 sequences conserved between human and pufferfish coincide with histone acetylation islands, and random sampling shows that 33% (13/39) of these can function as transcriptional enhancers in human Jurkat T cells. In addition, by comparing the human histone acetylation island sequences with mouse genome sequences, we find that despite the conservation of many of these regions between these species, 21,855 of these sequences are not conserved. Furthermore, we demonstrate that about 50% (26/51) of these nonconserved sequences have enhancer activity in Jurkat cells, and that many of the orthologous mouse sequences also have enhancer activity in addition to conserved epigenetic modification patterns in mouse T-cell chromatin. Therefore, by combining epigenetic modification and sequence data, we have established a novel genome-wide method for identifying regulatory elements not discernable by comparative genomics alone.
The sequencing of the genomes of an increasing number of organisms has enabled scientists to carry out comparative genomic analysis, and this has been particularly useful in the identification of evolutionarily conserved sequences, many of which have been shown to be important transcriptional regulatory elements (Hardison et al. 1997; Hardison 2000; Loots et al. 2000; Pennacchio and Rubin 2001; Miller et al. 2004). However, genomic comparisons based on DNA sequence alone have proved to be challenging due to various factors that can affect the outcome of a comparison, for instance, the alignment method used, the window size of the DNA sequences, and the stringency of other parameters used for comparison. Comparative genomics has revealed that a large number of conserved noncoding sequences exist between closely related species, thus making it difficult to identify sequences that are functionally significant. For this reason, genomic comparisons from multiple evolutionarily diverse species are often required to filter out regions with significant conservation, and even this can lead to the identification of false positives due to the presence of slowly evolving neutral regions (for review, see Stone et al. 2005). Comparative genomic analysis is also complicated due to degeneracy in functionally significant sequences, e.g., many transcription factors can recognize multiple binding sites and ultimately give rise to the same functional effect. In addition, conserved functional regions often have small amounts of neutrally evolving sequences embedded within them, and thus these regions may be missed in the analysis, particularly in more stringent comparisons.
The sequencing of the relatively small and compact genome of Fugu rubripes, the pufferfish, has provided us with a useful tool for the identification of conserved vertebrate functional elements (Brenner et al. 1993; Aparicio et al. 2002; Venkatesh and Yap 2004). This is due to the fact that the pufferfish has eliminated a large proportion of its nonessential DNA, yet this organism, which is one of the most distant vertebrate relatives of mammals, shares a similar repertoire of genes with mammals, implying that it uses similar regulatory mechanisms. However, it should be noted that a genome duplication occurs in pufferfish and other ray-finned fishes (Taylor et al. 2003; Christoffels et al. 2004), and it has been shown that some duplicated coparalogous regions have adapted new or subfunctional roles (e.g., Tümpel et al. 2006). Nevertheless, many conserved regulatory regions have been identified between pufferfish and mammals, e.g., the conserved regulatory elements that exist between the mouse and pufferfish Hoxb-4 gene (Aparicio et al. 1995). However, despite the fact that there is some regulatory conservation between these species, a large evolutionary distance still exists between them and, therefore, comparative genomics using the pufferfish genome would not likely identify the more specialized species-specific regulatory elements that have evolved to play pivotal roles in the more complex regulatory mechanisms of higher eukaryotes.
Studies of the regulation of eukaryotic gene expression have shown that not only the genomic DNA sequence per se, but also the higher order structure of chromatin plays an important role in the establishment and maintenance of tightly regulated gene expression. Many epigenetic modifications are known to occur in active and repressed chromatin regions (for review, see Jenuwein and Allis 2001; Felsenfeld and Groudine 2003; Peterson and Laniel 2004; Lam et al. 2005; Margueron et al. 2005; Martin and Zhang 2005), and many of these modifications are known to occur at important regulatory elements, e.g., the high levels of histone acetylation that are found at gene promoters and at many enhancers. Some of these modifications are known to be associated with active chromatin, including acetylation of histone H3 on lysines 9 and 14 and methylation on lysines 4, 36, and 79, while other modifications, including methylation of histone H3 on lysines 9 and 27, are linked with repressive chromatin environments, although recent data have shown an overlap of active and repressive modifications in distinct chromatin subpopulations (Bernstein et al. 2006; Roh et al. 2006). In addition, it is known that many regulatory elements carry these epigenetic modifications only in specific cell/tissue types or according to environmental conditions, which cannot be determined by comparative genomics based on sequence alone. Therefore, in carrying out genomic comparisons to identify regions that are likely to be functionally important, not only should the DNA sequence per se be taken into account, but it is also expected that the epigenetic modification patterns would be conserved in highly important regulatory regions. Such conservation of histone modifications has been demonstrated at several mammalian loci, e.g., conserved histone H3 acetylation has been shown at the locus control region (LCR) DNaseI hypersensitive site elements of avian and mammalian β-globin loci (Schübeler et al. 2000; Litt et al. 2001; Bulger et al. 2002, 2003). Epigenomic comparisons should thus be useful in filtering out functionally significant conserved noncoding sequences between closely related sequences, such as mouse and human. However, the major drawback in doing such comparisons has thus far been the lack of genome-wide information on epigenetic modification patterns in different species, in addition to the fact that these epigenetic modification patterns (but not the genome sequence) may vary from tissue to tissue and with changing environmental conditions depending on the chromatin dynamics of the gene(s) of interest.
In the present work, we make use of our previous study where we used a genome-wide mapping technique (GMAT) to determine the distribution of lysine-9/14-diacetylated histone H3 in human peripheral T cells (Roh et al. 2005). We have previously shown that this chromatin modification is correlated with active gene promoters and with important regulatory elements associated with gene expression, and we have defined regions of the genome carrying this modification as histone acetylation islands. Since these histone acetylation islands are very likely to denote functional elements, we compared their sequences with that of the pufferfish and mouse genomes, and random sampling with reporter gene assays in Jurkat cells revealed that a significant number of both conserved and nonconserved acetylation island sequences can function as enhancers. Moreover, we find that some orthologous regions in the mouse genome also display enhancer activity and have enriched histone acetylation patterns in mouse T-cell chromatin. Therefore, we show that by combining conventional comparative genomic strategies with genome-wide epigenetic information, we can more accurately pinpoint regions of functional significance as well as uncover functionally significant regions that are not necessarily conserved at the DNA sequence level, thus providing a novel genome-wide technique for identifying regulatory elements.
Results
We have previously mapped the acetylation of histone H3 in human peripheral T cells by using an unbiased genome-wide mapping technique we called GMAT (Roh et al. 2004, 2005, 2006). This technique, which is a combination of the chromatin immunoprecipitation (ChIP) assay and the serial analysis of gene expression (SAGE) assay, involves the isolation and subsequent sequencing of DNA tags from the acetylated histone-enriched chromatin, and the frequency of these tags in the sequenced DNA corresponds to the relative histone acetylation level at a particular location in the human T-cell genome. From this GMAT analysis, we identified 46,813 genomic regions where clusters of two or more unique acetylation tags are present, and we named these histone acetylation islands. Specifically, histone acetylation islands are defined based on the following criteria: (1) they are composed of GMAT sequence tags from more than two adjacent NlaIII sites (the restriction enzyme used to cleave the immunoprecipitated chromatin in the GMAT assay); (2) the detection frequency of the tags is ≥1; and (3) neighboring acetylation islands are separated by >500 bp. In addition, these acetylation island sequences ranged in length from 44 to 7827 bp (average size of 508 bp), and given that the resolution of the ChIP step of the GMAT assay was 500 bp–1 kb, the precise genomic location of the acetylated chromatin could be up to 1 kb from the location of the GMAT sequence tag. Many of these acetylation islands significantly correlated with conserved human–rodent noncoding sequences (CNSs) and with known regulatory elements in T cells, and high levels of H3 acetylation were detected in promoter regions, especially for genes that are highly expressed in human T cells (Roh et al. 2005).
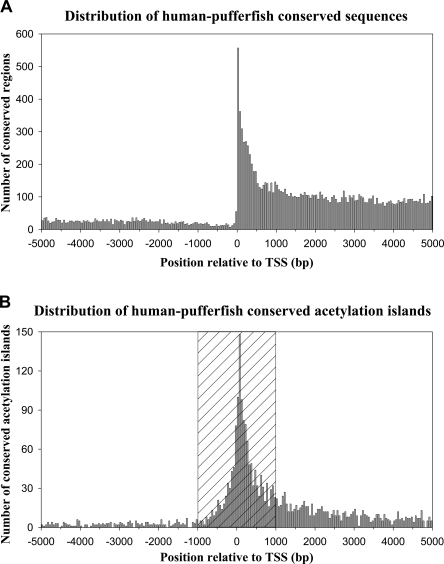
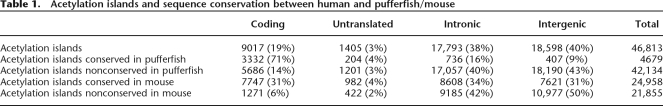
In the present study, to determine the extent of sequence conservation between the human T-cell histone acetylation islands and the pufferfish genome, we determined the precomputed aligned sequences of these genomes by using the VISTA genome browser (Mayor et al. 2000; Frazer et al. 2004; http://pipeline.lbl.gov/cgi-bin/gateway2). Of 91,915 genomic sequences that are conserved between these two distantly related vertebrates (of which 70,339 or 76.5% are also conserved in mouse, and 68,494 are protein-coding regions, 1338 are untranslated regions, and 22,083 are other noncoding sequences; see Methods for criteria used to define conservation), we found that 14,068 of these sequences (or 15%) are located within 5 kb of a RefSeq transcription start site (TSS) as downloaded from the UCSC Genome Browser (http://www.genome.ucsc.edu/). All human regions that showed a hit in the VISTA database were considered conserved irrespective of whether they occurred once or twice in the pufferfish genome. By counting the number of sequences in 50-bp window sizes, we plotted the distribution of 14,068 of these conserved sequences that are present within 5 kb of known or predicted TSSs in the human genome (Fig. 1A). We found that the majority of these conserved sequences tend to map downstream of a TSS or in the gene body rather than to an upstream or promoter region, which reflects the large number of conserved coding-region sequences. Upon examination of the distribution of conserved sequences that are associated with histone acetylation islands (see Table 1 for details), we found that 4679 acetylation islands coincide with conserved sequences and that 2459 of these fall within 5 kb of a TSS as plotted in Figure 1B. We found that a large portion (57.8%) of these conserved acetylation island sequences map within 1 kb of a TSS (hatched box, Fig. 1B). We also noted that many of these sequences are still present in the 5-kb region upstream of a TSS, suggesting that some of these may possibly be functional transcriptional regulatory elements in T cells, notwithstanding that many more distal regulatory elements associated with histone acetylation may exist beyond the 5-kb limit examined here.
Figure 1.
The distribution of sequences that are conserved between human and pufferfish. (A) The distribution of human–pufferfish conserved sequences. A total of 14,068 conserved sequences are plotted relative to their location within 5 kb of the nearest RefSeq transcription start site (TSS) in the human genome. The y-axis indicates the number of sequences (50-bp window sizes) found 5 kb upstream or downstream of a TSS. (B) The distribution of human–pufferfish conserved sequences that are associated with histone acetylation islands. A total of 2459 conserved sequences associated with acetylation islands are plotted as in A. The hatched box indicates sequences found within 1 kb of a TSS.
Table 1.
Acetylation islands and sequence conservation between human and pufferfish/mouse
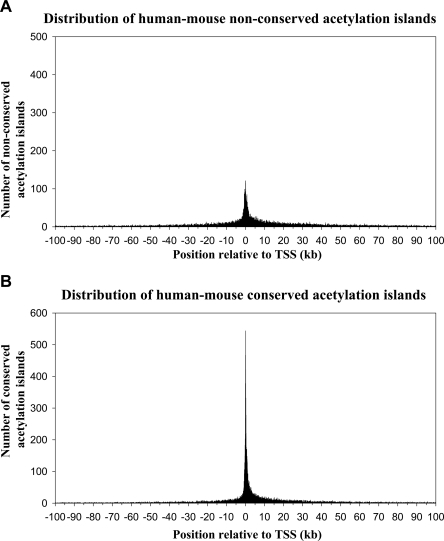
In order to examine the extent of sequence conservation of the human T-cell histone acetylation islands with a more closely related organism, we next compared these acetylation island sequences with mouse genome sequences. In total, we found that 1,362,767 genomic regions are conserved between human and mouse by VISTA analysis, of which 162,613 are protein-coding regions, 37,664 are untranslated regions, and 1,162,490 are other noncoding sequences. By comparing our 46,813 human T-cell acetylation island sequences with these conserved sequences, we found that 24,958 acetylation islands coincide with conserved regions (see Table 1 for details). In addition, we were also interested in using our histone acetylation island data to uncover sequences that are not conserved between human and mouse, but which may nonetheless represent functional elements. From this analysis, we calculated the relative distance of 19,070 nonconserved acetylation island sequences from the nearest RefSeq TSS in the human genome, and by counting the number of islands in 50-bp window sizes, we plotted their distribution within 100 kb of the TSSs (Fig. 2A). Again, as in Figure 1, we find that the majority of these nonconserved sequences map at or near a TSS. The fact that a significant portion of these acetylation island sequences are present at large distances (>20 kb) from a TSS suggests that they may possibly represent distant functional elements involved in long-range gene regulation or chromatin domain formation such as enhancers, LCRs, matrix attachment regions, or boundary elements, or they could possibly represent unknown noncoding RNA genes and/or intergenic transcripts. We also wanted to compare the distribution of these nonconserved acetylation island sequences to that of conserved acetylation island sequences. We therefore plotted the distribution of 22,796 of these conserved sequences that fall within 100 kb of a TSS (Fig. 2B). This distribution of conserved acetylation island sequences was not surprisingly similar to that of the nonconserved sequences, but we noted that a larger number of the conserved sequences map at or close to a TSS.
Figure 2.
The distribution of human–mouse conserved and nonconserved histone acetylation island sequences. (A) The distribution of 19,070 nonconserved acetylation island sequences is plotted relative to their location within 100 kb of the nearest RefSeq TSS. The y-axis indicates the number of sequences (50-bp window sizes) found ±100 kb from a TSS. (B) The distribution of 22,796 human–mouse conserved acetylation island sequences is plotted as in A.
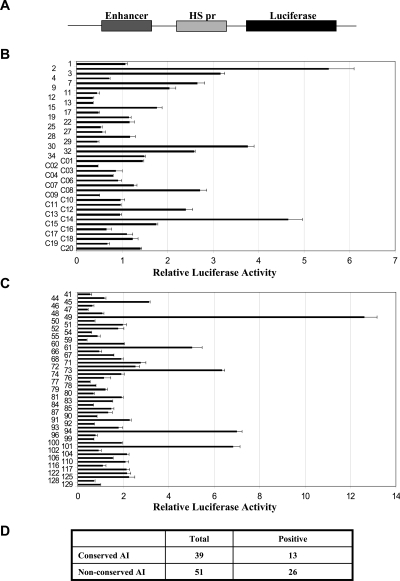
To determine what proportion of the human–pufferfish conserved, or human–mouse nonconserved, acetylation island sequences display functional enhancer activity, we decided to randomly sample some of these human sequences for enhancer activity in reporter gene assays. In these assays, we inserted 1.2-kb test sequences spanning the acetylation islands upstream of a heat-shock promoter-driven luciferase reporter gene (Fig. 3A), and we transiently transfected these constructs into human Jurkat T cells. By comparing the luciferase activity of each construct to the same construct without an insertion, we determined the relative enhancer activities of the selected acetylation island sequences (Fig. 3B,C; Supplemental Tables S1 and S2). To test for enhancer activity of the human–pufferfish conserved sequences, 39 sequences located >1 kb from a TSS were selected, omitting those sequences in the hatched box in Figure 1. The data show that of the 39 human conserved sequences tested, 13 show a minimum of 1.5-fold enhancer activity (Fig. 3 B,D; Supplemental Table S1), consistent with the expectation that these sequences represent functional elements. In addition, we tested 51 human–mouse nonconserved sequences that are located >1 kb from a transcription start site, and we found that 26 of these displayed enhancer activity in our assay (Fig. 3C,D; Supplemental Table S2), suggesting that these histone acetylation islands are important elements in human T cells, despite the lack of sequence conservation. Given that surprisingly high numbers of our conserved and nonconserved sequences showed enhancer activity in our assay, 33% and 50%, respectively, it was therefore necessary to determine whether the enhancer activity seen in these sequences was statistically significant for acetylated regions. We therefore randomly chose nine nonacetylated sequences in the human genome, and upon testing these in our reporter gene assay, we found no significant enhancer activity above our threshold level (Supplemental Table S3), strongly suggesting that a significant portion of our sampled acetylated sequences are bone fide regulatory elements. Furthermore, by utilizing the results with these randomly chosen nonacetylated sequences, we performed Student t-tests and calculated P-values for all acetyl-associated sequences that showed enhancer activity in our assays (Fig. 3; Supplemental Tables S1 and S2), and the very low P-values obtained, ranging from 8.1 × 10−13 to 1.3 × 10−5 (Supplemental Tables S1 and S2), revealed that our enhancer activity is statistically significant.
Figure 3.
Enhancer activity assays of the conserved and nonconserved acetylation island sequences. (A) The construct used to test for enhancer activity. Each construct contains a 1.2-kb sequence encompassing the acetylation island sequence, which is cloned upstream of a luciferase reporter gene driven by a heat-shock gene promoter (HS pr). (B) Reporter gene activities of 39 acetylation island sequences that are conserved between human and pufferfish, following transient transfection of the constructs in Jurkat cells. The relative luciferase activity (x-axis) was determined by comparing the luciferase activity of the test construct to that of a similar construct without a putative enhancer insertion. The construct numbers (as shown in Supplemental Table S1) are indicated on the y-axis. Error bars indicate the standard deviation from the mean. (C) Reporter gene activities of 51 acetylation island sequences that are not conserved between human and mouse, as plotted in B. The construct numbers on the y-axis are described in Supplemental Table S2. (D) Summary of enhancer assay results. (AI) Acetylation island.
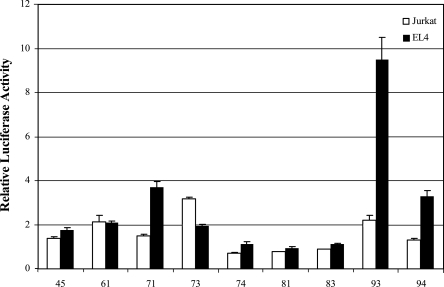
Given the distribution of the human nonconserved acetylation island sequences (Fig. 2A) and the fact that about half of these displayed enhancer activity (Fig. 3C,D), we were interested in determining whether the orthologous nonconserved mouse sequences were functional in enhancer assays and whether or not they too had acetylated histone H3 in mouse T cells. We thus selected nine of these nonconserved human acetylation island sequences that showed enhancer activity in the above assays, and by using the VISTA genome browser, we determined the spatially equivalent or candidate orthologous sequences in the mouse genome (Supplemental Table S4), which, according to the criteria used by the browser, correspond to sequences that are located between conserved sequence blocks in syntenic human–mouse genomic regions. We then tested these sequences in enhancer assays (Supplemental Table S4), comparing their enhancer activities in human Jurkat T cells to that in the mouse thymoma-derived EL4 T-cell line (Fig. 4). Of the sequences tested in Jurkat cells (Fig. 4; Supplemental Table S4), we found that 44% of the corresponding mouse sequences displayed enhancer activity, suggesting that although the sequence per se is not conserved, there is still some functional conservation between these corresponding regions. Furthermore, when these sequences were tested for enhancer activity in mouse EL4 cells, which do not have the cross-species variations that may be present in human Jurkat cells, we found that an even higher proportion of these sequences, 67%, displayed enhancer activity (Fig. 4; Supplemental Table S4).
Figure 4.
Enhancer activities of nonconserved but corresponding mouse sequences. The enhancer activities of nine nonconserved orthologous mouse sequences are shown following transient transfection in human Jurkat cells and mouse EL4 cells. Relative enhancer activities were determined as in Figure 3. The construct numbers (x-axis) are described in Supplemental Table S4.
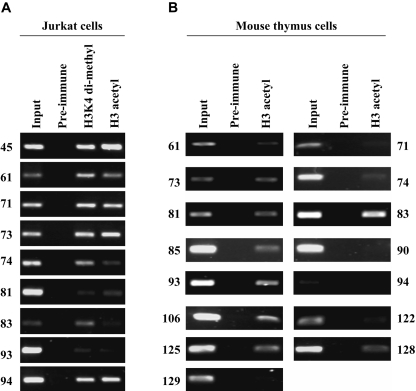
We next wanted to determine the histone acetylation modification patterns of these sequences in Jurkat cells and in mouse T-cell chromatin. We first did ChIP assays with some of the human sequences that displayed enhancer activity in Jurkat cells (Fig. 5A). Although these sequences were defined as histone acetylation islands in the GMAT analysis in human peripheral pan T cells (Roh et al. 2005), the Jurkat cells used in the enhancer assays are a CD4+ T-cell line and may not have the same modifications as the primary T cells used in the GMAT analysis. We therefore analyzed both lysine 9/14 diacetylation of histone H3 and lysine 4 dimethylation of histone H3 (Fig. 5A), and we found that most (at least seven of the nine sequences tested), but not all, of these sequences displayed these histone modifications in Jurkat cells. We next wanted to sample orthologous mouse sequences to see whether they also have conserved histone acetylation in mouse T-cell chromatin, similar to their counterparts being histone acetylation islands in human T-cell chromatin. We thus performed ChIP assays with a lysine 9/14 diacetyl histone H3 antibody on 15 different orthologous mouse regions, and we found that about two-thirds of these sequences, which are not conserved at the sequence level, also carry this histone modification in mouse thymocytes (Fig. 5B). Therefore, both the enhancer assays (Fig. 4) and the histone modification analysis (Fig. 5) may possibly imply that at least some of these nonconserved but corresponding mouse sequences have functional conservation (notwithstanding possible functional changes due to adaptive evolution or neofunctionalization) as determined not by sequence per se, but by epigenetic modification patterns.
Figure 5.
Histone modifications of the acetylation island sequences. (A) ChIP assays using diacetyl K9/K14 histone H3 and dimethyl K4 histone H3 antibodies in human Jurkat cells. The sequence numbers are described in Supplemental Table S2. (B) ChIP assays using a diacetyl K9/K14 histone H3 antibody in mouse thymus cells. The sequence numbers are described in Supplemental Tables S2 and S4.
Discussion
In this study, we have taken advantage of our previous genome-wide histone acetylation data in human T cells, and we have compared acetylation island sequences with the genomic sequences of a distantly related vertebrate, the pufferfish, as well as to a more closely related species, the mouse. In this way, we have identified many evolutionarily conserved sequences between pufferfish and human, and we have shown that about one-third of these sequences act as enhancers in reporter gene assays, and these sequences are thus likely to be highly functional. In the case of our human–mouse acetylation island sequence comparisons, we have focused on sequences that are not conserved between these species. Our data reveal that about half of these sequences have enhancer activity in human Jurkat T cells, and thus, are likely to represent important regulatory regions. Furthermore, we have shown that about two-thirds of the sampled orthologous but nonconserved mouse sequences show enhancer activity in our mouse T-cell line reporter gene assays, and that many of these sequences also have conserved histone acetylation in mouse thymocytes. Therefore, given that 40% of the mouse genome can be aligned with the human genome (Waterston et al. 2002), which complicates narrowing down the functionally important conserved regions, our use of epigenomic data to identify genome-wide regulatory sequences, including those that are not conserved, represents a step forward in the comparative genomics field.
While surprisingly large numbers of both our conserved and nonconserved acetylation island sequences function as enhancers in our reporter gene assays, we cannot exclude the possibility that those that did not show enhancer activity may still have functional significance. These histone acetylation island sequences could possibly have enhancer activity in other cell lines, at different developmental stages, in different environmental conditions, in combination with other elements in the chromatin domains they occupy, or they may not be compatible with the heat-shock promoter we tested in our assays. It is also possible that these histone-acetylated regions are not enhancers but may represent other regions of biological importance, e.g., they could be boundary elements, sites of DNA recombination or repair, or replication origins. It is known that high levels of histone acetylation are not necessarily restricted to promoter or enhancer regions, and there are various reports in the literature of such instances (e.g., Ikura et al. 2000; McBlane and Boyes 2000; McMurry and Krangel 2000; Litt et al. 2001; Bird et al. 2002; Mutskov et al. 2002; Bulger et al. 2003; Aggarwal and Calvi 2004). In addition, in the case of the conserved and nonconserved sequences that showed enhancer activity in our assays, it is important to note that we cannot make conclusions about functionality based on these assays alone, even when there is conservation of the histone acetylation islands, and various speculations can be made about these regions, including the possibility of functional changes due to adaptive evolution or neofunctionalization, or the possibility that they could represent unknown noncoding RNAs.
In the case of the nonconserved orthologous regions in the mouse genome that did not show enhancer activity, it is possible that the human T-cell acetylation islands could represent species-specific enhancer/regulatory regions. An example of this is the nonconserved acetylation island sequence that is present downstream of the IL13 gene in the TH2 cytokine locus (Roh et al. 2005), which juxtaposes another acetylation island previously identified as a CNS that is required for the coordinated expression of the TH2 genes (Loots et al. 2000). However, in analyzing the nonconserved but “corresponding” mouse sequences, it is important to note that the accuracy of defining these highly diverged but orthologous regions is limited by the capabilities of the VISTA alignment program we used. While this aligner is generally used to compute global alignments and is constantly being improved to handle chromosomal rearrangements like inversions, deletions, duplications, insertions, and nucleotide substitutions (Mayor et al. 2000; Couronne et al. 2003; Frazer et al. 2003, 2004), there is always the possibility that the precomputed aligned sequences are not truly orthologous or corresponding regions, and perhaps another alignment method, such as a local alignment program or phylogenetic footprint analyses, could uncover more precise corresponding regions or determine whether, at all, a truly orthologous region exists in the other species. For this reason, to determine the biological/evolutionary relevance of the individual nonconserved acetylated sequences tested here, further in-depth analyses of our epigenomic comparisons would have to involve more precise local alignments and factor binding-site data, which is beyond the scope of the current study. Nonetheless, despite these limitations, it is still remarkable that such a high proportion of our nonconserved sequences showed enhancer activity in our assays, and that many of these had conserved histone acetylation in mouse T-cell chromatin. Similar examples of conserved histone modifications in nonconserved human and mouse sequences were also shown in a study by Bernstein et al. (2005), where a comparison of histone H3 lysine 4 methylation was carried out across human chromosomes 21 and 22 and at a series of human and mouse loci. However, our study has been extended to a genome-wide scale, and we have carried the analyses a step further by demonstrating in vitro enhancer activity at nonconserved human–mouse sequences in addition to showing conserved histone acetylation at these regions.
While our study is part of a new era in epigenomic sequence comparisons, the concept of using epigenetic features to identify important chromatin regulatory regions has been used in biology for some time now, e.g., the occurrence of CpG islands and their methylation status, or the occurrence of DNaseI hypersensitive sites that were later shown to have enhancer, LCR, or boundary element activity, even before the underlying DNA sequences were known. However, most of these previous studies were limited by the availability of sequences and/or clones, and could only be confined to the specific loci under investigation. Nowadays, the recent sequencing of the genomes of a growing number of higher eukaryotic organisms, coupled with the emergence of new technologies to map chromatin modifications genome-wide (for review, see Huebert and Bernstein 2005; Barrera and Ren 2006; Callinan and Feinberg 2006), including our GMAT analysis, has paved the way for future breakthroughs in deciphering the functionally relevant areas of the human genome. Not only can histone modification data be used for such purposes, but knowledge of the genome-wide binding sites of specific transcription factors, or of DNA methylation patterns, or of DNaseI hypersensitivity, could also be applied to identify functionally significant genomic regions. To this end, a recent study has used genome-wide chromatin immunoprecipitation studies to identify promoters in the human genome independently of the mRNA sequences (Kim et al. 2005a, b). In addition, methodologies have been developed to map DNaseI hypersensitivity genome-wide (Dorschner et al. 2004; Sabo et al. 2004; Crawford et al. 2006), and recent studies have also compared DNA methylation in normal versus human cancer cells (Weber et al. 2005; Callinan and Feinberg 2006; Wilson et al. 2006).
In conclusion, we have combined genome-wide epigenomic data with conventional comparative genomics to reveal both conserved and nonconserved functionally significant regions in the human and mouse genomes, and we have thereby uncovered new functional regions that were not discernable by sequence comparisons alone. In the future, this method could be extended to yield even more information by comparing our human T-cell histone modified sequences to genome-wide histone modifications in mouse T-cell chromatin. In the continuing efforts to mine the human genome and to weed out significant regulatory regions, it has become clear that genomic comparisons between different organisms have their limitations, and future comparisons must not only take into account the DNA sequence itself but also the architecture of the surrounding chromatin and its associated modifications. The emergence of future genome-wide epigenomic data from various cell types and organisms will undoubtedly aid in these epigenomic comparisons and thus greatly increase our chances of finding important regulatory regions, which in turn will further our understanding of gene regulation, developmental processes, and the underlying causes of human diseases.
Methods
Sequence alignments
For sequence comparisons, the UCSC human July 2003 (hg16/NCBI build 34) base sequence assembly was compared with the mouse mm4 October 2003 assembly or to the Fugu v.3.0 August 2002 assembly. Conserved and corresponding aligned sequences were downloaded based on precomputed alignments from the VISTA analysis Web site (http://genome.lbl.gov/vista/index.shtml) using the default sliding window size of 100 bp to calculate conservation scores for each base pair in the VISTA curve. Regions that had a minimum of 70% identity were considered conserved in our analysis, and all other parameters were as defined for the precomputed alignments in the VISTA database. The RefSeq gene tables of the human hg16 assembly were downloaded from the UCSC Web site (http://www.genome.ucsc.edu/). Histone acetylation island sequences were obtained as described previously (Roh et al. 2005; http://dir.nhlbi.nih.gov/labs/lmi/zhao/epigenome/G&D2005.htm).
Cell lines
Jurkat cells and EL4 cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum and 1% penicillin–streptomycin mix.
Reporter gene assays
Enhancer candidates were amplified using High Fidelity Platinum Taq DNA polymerase (Invitrogen), and all products were about 1.2 kb in length with the acetylation island sequences (Supplemental Tables S1–S4) in the center. Primer sequences are available upon request. To test these sequences in enhancer assays, pGL3-HS was constructed by subcloning the SmaI–NheI (blunt-ended) minimal heat-shock promoter fragment from pIND (Invitrogen) into the blunt-ended HindIII site of pGL3-basic (Promega). The amplified candidate sequences were then subcloned in either the KpnI or BglII restriction sites of the pGL3-HS construct immediately upstream of the heat-shock promoter. Jurkat cells were transfected with 2 μg of each reporter construct using Superfect transfecting reagent (Qiagen) according to the manufacturer’s directions, and EL4 cells were transfected with 10 μg of each reporter construct by electroporation with a BIO-RAD gene pulser (250 V, 960 μF). Cell extracts were prepared, and luciferase activities were measured 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega). All enhancer assays were carried out at least in duplicate.
ChIP analysis
ChIP assays using Jurkat cells and mouse thymocytes were performed as described previously (Roh et al. 2004). Pre-immune serum (Santa Cruz Biotechnologies), antidiacetylated K9/K14 histone H3 (Upstate) and dimethyl K4 histone H3 (Abcam) antibodies were used for immunoprecipitation. Primer sequences used in PCR reactions are available upon request. Input and immunoprecipitated DNA samples were examined by agarose gel electrophoresis after 32 cycles of PCR amplification.
Acknowledgments
We thank Drs. Michael Zhang and Warren Leonard for critical reading of the manuscript, Dr. Dangsheng Li and members of the Zhao laboratory for helpful suggestions and discussion, and Dr. Deyou Zheng for suggesting the examination of regions 25 and 34 in Supplemental Table S1. This work was supported by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5767907
References
- Aggarwal B.D., Calvi B.R., Calvi B.R. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- Aparicio S., Morrison A., Gould A., Gilthorpe J., Chaudhuri C., Rigby P., Krumlauf R., Brenner S., Morrison A., Gould A., Gilthorpe J., Chaudhuri C., Rigby P., Krumlauf R., Brenner S., Gould A., Gilthorpe J., Chaudhuri C., Rigby P., Krumlauf R., Brenner S., Gilthorpe J., Chaudhuri C., Rigby P., Krumlauf R., Brenner S., Chaudhuri C., Rigby P., Krumlauf R., Brenner S., Rigby P., Krumlauf R., Brenner S., Krumlauf R., Brenner S., Brenner S. Detecting conserved regulatory elements with the model genome of the Japanese pufferfish, Fugu rubripes. Proc. Natl. Acad. Sci. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S., Chapman J., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chapman J., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Christoffels A., Rash S., Hoon S., Smit A., Rash S., Hoon S., Smit A., Hoon S., Smit A., Smit A., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Barrera L.O., Ren B., Ren B. The transcriptional regulatory code of eukaryotic cells–insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr. Opin. Cell Biol. 2006;18:1–8. doi: 10.1016/j.ceb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., McMahon S., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Karlsson E.K., Kulbokas E.J., III, Gingeras T.R., Kulbokas E.J., III, Gingeras T.R., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Fry B., Meissner A., Wernig M., Plath K., Meissner A., Wernig M., Plath K., Wernig M., Plath K., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bird A.W., Yu D.Y., Pray-Grant M.G., Qiu Q., Harmon K.E., Megee P.C., Grant P.A., Smith M.M., Christman M.F., Yu D.Y., Pray-Grant M.G., Qiu Q., Harmon K.E., Megee P.C., Grant P.A., Smith M.M., Christman M.F., Pray-Grant M.G., Qiu Q., Harmon K.E., Megee P.C., Grant P.A., Smith M.M., Christman M.F., Qiu Q., Harmon K.E., Megee P.C., Grant P.A., Smith M.M., Christman M.F., Harmon K.E., Megee P.C., Grant P.A., Smith M.M., Christman M.F., Megee P.C., Grant P.A., Smith M.M., Christman M.F., Grant P.A., Smith M.M., Christman M.F., Smith M.M., Christman M.F., Christman M.F. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- Brenner S., Elgar G., Sandford R., Macrae A., Venkatesh B., Aparicio S., Elgar G., Sandford R., Macrae A., Venkatesh B., Aparicio S., Sandford R., Macrae A., Venkatesh B., Aparicio S., Macrae A., Venkatesh B., Aparicio S., Venkatesh B., Aparicio S., Aparicio S. Characterization of the pufferfish (fugu) genome as a compact model vertebrate genome. Nature. 1993;306:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- Bulger M., Sawado T., Schübeler D., Groudine M., Sawado T., Schübeler D., Groudine M., Schübeler D., Groudine M., Groudine M. ChIPs of the β-globin locus: Unraveling gene regulation within an active domain. Curr. Opin. Genet. Dev. 2002;12:170–177. doi: 10.1016/s0959-437x(02)00283-6. [DOI] [PubMed] [Google Scholar]
- Bulger M., Schübeler D., Bender M.A., Hamilton J., Farrell C.M., Hardison R.C., Groudine M., Schübeler D., Bender M.A., Hamilton J., Farrell C.M., Hardison R.C., Groudine M., Bender M.A., Hamilton J., Farrell C.M., Hardison R.C., Groudine M., Hamilton J., Farrell C.M., Hardison R.C., Groudine M., Farrell C.M., Hardison R.C., Groudine M., Hardison R.C., Groudine M., Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan P.A., Feinberg A.P., Feinberg A.P. The emerging science of epigenomics. Hum. Mol. Genet. 2006;15:R95–R101. doi: 10.1093/hmg/ddl095. [DOI] [PubMed] [Google Scholar]
- Christoffels A., Koh E.G., Chia J.M., Brenner S., Aparicio S., Venkatesh B., Koh E.G., Chia J.M., Brenner S., Aparicio S., Venkatesh B., Chia J.M., Brenner S., Aparicio S., Venkatesh B., Brenner S., Aparicio S., Venkatesh B., Aparicio S., Venkatesh B., Venkatesh B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol. 2004;21:1146–1151. doi: 10.1093/molbev/msh114. [DOI] [PubMed] [Google Scholar]
- Couronne O., Poliakov A., Bray M., Ishkhanov T., Ryaboy D., Rubin E., Pachter L., Dubchak I., Poliakov A., Bray M., Ishkhanov T., Ryaboy D., Rubin E., Pachter L., Dubchak I., Bray M., Ishkhanov T., Ryaboy D., Rubin E., Pachter L., Dubchak I., Ishkhanov T., Ryaboy D., Rubin E., Pachter L., Dubchak I., Ryaboy D., Rubin E., Pachter L., Dubchak I., Rubin E., Pachter L., Dubchak I., Pachter L., Dubchak I., Dubchak I. Strategies and tools for whole-genome alignments. Genome Res. 2003;13:73–80. doi: 10.1101/gr.762503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford G.E., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., Chen Y., Bernat J.A., Ginsburg D., Bernat J.A., Ginsburg D., Ginsburg D., et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner M.O., Hawrylycz M., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Hawrylycz M., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Mack J., Hall R., Goldy J., Sabo P.J., Hall R., Goldy J., Sabo P.J., Goldy J., Sabo P.J., Sabo P.J., et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nat. Methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., Groudine M., Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Frazer K.A., Elnitski L., Church D.M., Dubchak I., Hardison R.C., Elnitski L., Church D.M., Dubchak I., Hardison R.C., Church D.M., Dubchak I., Hardison R.C., Dubchak I., Hardison R.C., Hardison R.C. Cross-species sequence comparisons: A review of methods and available resources. Genome Res. 2003;13:1–12. doi: 10.1101/gr.222003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I., Pachter L., Poliakov A., Rubin E.M., Dubchak I., Poliakov A., Rubin E.M., Dubchak I., Rubin E.M., Dubchak I., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R.C. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Hardison R., Slightom J.L., Gumucio D.L., Goodman M., Stojanovic N., Miller W., Slightom J.L., Gumucio D.L., Goodman M., Stojanovic N., Miller W., Gumucio D.L., Goodman M., Stojanovic N., Miller W., Goodman M., Stojanovic N., Miller W., Stojanovic N., Miller W., Miller W. Locus control regions of mammalian β-globin gene clusters: Combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- Huebert D.J., Bernstein B.E., Bernstein B.E. Genomic views of chromatin. Curr. Opin. Genet. Dev. 2005;15:476–481. doi: 10.1016/j.gde.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Ikura T., Ogryzko V.V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y., Ogryzko V.V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y., Horikoshi M., Scully R., Qin J., Nakatani Y., Scully R., Qin J., Nakatani Y., Qin J., Nakatani Y., Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Qu C., Van Calcar S., Trinklein N.D., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld J.G., Myers R.M., Barrera L.O., Qu C., Van Calcar S., Trinklein N.D., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld J.G., Myers R.M., Qu C., Van Calcar S., Trinklein N.D., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld J.G., Myers R.M., Van Calcar S., Trinklein N.D., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld J.G., Myers R.M., Trinklein N.D., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld J.G., Myers R.M., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld J.G., Myers R.M., Luna R.M., Glass C.K., Rosenfeld J.G., Myers R.M., Glass C.K., Rosenfeld J.G., Myers R.M., Rosenfeld J.G., Myers R.M., Myers R.M., et al. Direct isolation and identification of promoters in the human genome. Genome Res. 2005a;15:830–839. doi: 10.1101/gr.3430605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005b;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam A.L., Pazin D.E., Sullivan B.A., Pazin D.E., Sullivan B.A., Sullivan B.A. Control of gene expression and assembly of chromosomal subdomains by chromatin regulators with antagonistic functions. Chromosoma. 2005;114:242–251. doi: 10.1007/s00412-005-0001-0. [DOI] [PubMed] [Google Scholar]
- Litt M.D., Simpson M., Recillas-Targa F., Prioleau M.N., Felsenfeld G., Simpson M., Recillas-Targa F., Prioleau M.N., Felsenfeld G., Recillas-Targa F., Prioleau M.N., Felsenfeld G., Prioleau M.N., Felsenfeld G., Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots G.G., Locksley R.M., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Locksley R.M., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Miller W., Rubin E.M., Frazer K.A., Rubin E.M., Frazer K.A., Frazer K.A. Identification of a coordinate regulator of interleukins 4, 13 and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Margueron R., Trojer P., Reinberg D., Trojer P., Reinberg D., Reinberg D. The key to development: Interpreting the histone code? Curr. Opin. Genet. Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Martin C., Zhang Y., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Mayor C., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I., Frazer K.A., Pachter L.S., Dubchak I., Pachter L.S., Dubchak I., Dubchak I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- McBlane F., Boyes J., Boyes J. Stimulation of V(D)J recombination by histone acetylation. Curr. Biol. 2000;10:483–486. doi: 10.1016/s0960-9822(00)00449-8. [DOI] [PubMed] [Google Scholar]
- McMurry M.T., Krangel M.S., Krangel M.S. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- Miller W., Makova K.D., Nekrutenko A., Hardison R.C., Makova K.D., Nekrutenko A., Hardison R.C., Nekrutenko A., Hardison R.C., Hardison R.C. Comparative genomics. Annu. Rev. Genomics Hum. Genet. 2004;5:15–56. doi: 10.1146/annurev.genom.5.061903.180057. [DOI] [PubMed] [Google Scholar]
- Mutskov V.J., Farrell C.M., Wade P.A., Wolffe A.P., Felsenfeld G., Farrell C.M., Wade P.A., Wolffe A.P., Felsenfeld G., Wade P.A., Wolffe A.P., Felsenfeld G., Wolffe A.P., Felsenfeld G., Felsenfeld G. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes & Dev. 2002;16:1540–1554. doi: 10.1101/gad.988502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A., Rubin E.M., Rubin E.M. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Peterson C.L., Laniel M.A., Laniel M.A. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Roh T.Y., Ngau W.C., Cui K., Landsman D., Zhao K., Ngau W.C., Cui K., Landsman D., Zhao K., Cui K., Landsman D., Zhao K., Landsman D., Zhao K., Zhao K. High-resolution genome-wide mapping of histone modifications. Nat. Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Zhao K., Cuddapah S., Zhao K., Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes & Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Cui K., Zhao K., Cuddapah S., Cui K., Zhao K., Cui K., Zhao K., Zhao K. The landscape of histone modifications in human T Cells. Proc. Natl. Acad. Sci. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo P.J., Hawrylycz M., Wallace J.C., Humbert R., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Hawrylycz M., Wallace J.C., Humbert R., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Wallace J.C., Humbert R., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Humbert R., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., Kawamoto J., Hall R., Mack J., Dorschner M.O., Hall R., Mack J., Dorschner M.O., Mack J., Dorschner M.O., Dorschner M.O., et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc. Natl. Acad. Sci. 2004;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D., Francastel C., Cimbora D.M., Reik A., Martin D.I., Groudine M., Francastel C., Cimbora D.M., Reik A., Martin D.I., Groudine M., Cimbora D.M., Reik A., Martin D.I., Groudine M., Reik A., Martin D.I., Groudine M., Martin D.I., Groudine M., Groudine M. Nuclear localization and histone acetylation: A pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes & Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- Stone E.A., Cooper G.M., Sidow A., Cooper G.M., Sidow A., Sidow A. Trade-offs in detecting evolutionarily constrained sequence by comparative genomics. Annu. Rev. Genomics Hum. Genet. 2005;6:143–164. doi: 10.1146/annurev.genom.6.080604.162146. [DOI] [PubMed] [Google Scholar]
- Taylor J.S., Braasch I., Frickey T., Meyer A., de Van Peer Y., Braasch I., Frickey T., Meyer A., de Van Peer Y., Frickey T., Meyer A., de Van Peer Y., Meyer A., de Van Peer Y., de Van Peer Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tümpel S., Cambronero F., Wiedemann L.M., Krumlauf R., Cambronero F., Wiedemann L.M., Krumlauf R., Wiedemann L.M., Krumlauf R., Krumlauf R. Evolution of cis elements in the differential expression of two Hoxa2coparologous genes in pufferfish (Takifugu rubripes) Proc. Natl. Acad. Sci. 2006;103:5419–5424. doi: 10.1073/pnas.0600993103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B., Yap W.H., Yap W.H. Comparative genomics using fugu: A tool for the identification of conserved vertebrate cis-regulatory elements. Bioessays. 2004;27:100–107. doi: 10.1002/bies.20134. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schübeler D., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schübeler D., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schübeler D., Oakeley E.J., Haase M., Lam W.L., Schübeler D., Haase M., Lam W.L., Schübeler D., Lam W.L., Schübeler D., Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Wilson I.M., Davies J.J., Weber M., Brown C.J., Alvarez C.E., MacAulay C., Schübeler D., Lam W.L., Davies J.J., Weber M., Brown C.J., Alvarez C.E., MacAulay C., Schübeler D., Lam W.L., Weber M., Brown C.J., Alvarez C.E., MacAulay C., Schübeler D., Lam W.L., Brown C.J., Alvarez C.E., MacAulay C., Schübeler D., Lam W.L., Alvarez C.E., MacAulay C., Schübeler D., Lam W.L., MacAulay C., Schübeler D., Lam W.L., Schübeler D., Lam W.L., Lam W.L. Epigenomics: Mapping the methylome. Cell Cycle. 2006;5:155–158. doi: 10.4161/cc.5.2.2367. [DOI] [PubMed] [Google Scholar]