Abstract
HepSEQ is a repository for an extensive library of public health and molecular data relating to hepatitis B virus (HBV) infection collected from international sources. It is hosted by the Centre for Infections, Health Protection Agency (HPA), England, United Kingdom. This repository has been developed as a web-enabled, quality-controlled database to act as a tool for surveillance, HBV case management and for research. The web front-end for the database system can be accessed from http://www.hpa-bioinfodatabases.org.uk/hepatitis_open/main.php. The format of the database system allows for comprehensive molecular, clinical and epidemiological data to be deposited into a functional database, to search and manipulate the stored data and to extract and visualize the information on epidemiological, virological, clinical, nucleotide sequence and mutational aspects of HBV infection through web front-end. Specific tools, built into the database, can be utilized to analyse deposited data and provide information on HBV genotype, identify mutations with known clinical significance (e.g. vaccine escape, precore and antiviral-resistant mutations) and carry out sequence homology searches against other deposited strains. Further mechanisms are also in place to allow specific tailored searches of the database to be undertaken.
INTRODUCTION
Viral hepatitis due to hepatitis B virus (HBV) is a major worldwide public health concern leading to acute and chronic liver disease including cirrhosis and hepatocellular carcinoma (HCC) (1–4). It is currently estimated that over 2.5 billion people are exposed and over 350 million people are chronically infected with the virus and that ∼1.2 million people die annually from HBV-related disease (5–7). The prevalence of HBV is known to be higher through Asia and the Middle East, Africa, South America and the Mediterranean countries. In these regions, transmission occurs mainly through vertical and horizontal routes. In North America and Northern Europe, where HBV prevalence is lower, sexual and intravenous drug use are the major modes of transmission (8,9).
There are few reliable predicators for the risk of developing serious consequences of HBV infection such as host-related factors (gender, age at infection, degree of liver damage at presentation and immune competence), environmental factors (alcohol consumption, co-infection with other viruses such as HIV and HCV and drug therapy) and HBV-related factors (serological markers, viral load and persistence of viral replication). HBV is currently classified into eight genotypes (A–H) based on sequence divergence over the entire genome exceeding 8% at the nucleotide level (10–12). These major genotypes have a distinct geographical distribution (13,14). Additional variability in the genome has been shown to arise as a result of the natural emergence of strains which may have a selective advantage during the course of chronic HBV infection in a patient, e.g. precore mutants, deletions in the core gene, preS1 and preS2 regions [for a review see (15)]. It is speculated that these variants are driven by the immune system but it currently remains unknown which, if any are clinically significant. Sequence evolution driven by external pressures such as the introduction of immunization programmes and more recently antiviral treatment has also given rise to a number of mutations within the viral polymerase and envelope regions [for a review see (16)].
Although in vitro studies have provided significant information into understanding the clinical significance of sequence changes, these data remain limited to a few specific mutations (17,18). The development of databases containing detailed genetic sequences of human pathogens provides a new point of departure for the investigation of host–parasite relationships. Using bioinformatics techniques it is possible to assess pathogen relatedness and likely evolutionary pathways, and to examine the pathways of sequence evolution of an agent in response to a particular selection pressure such as antiviral treatment. Furthermore, building a repository for such data allows for the monitoring of the distribution and variability of HBV strains at regional, national and global levels, which is of importance in an increasingly mobile population. Furthermore, such data provide a powerful tool in the public health setting when investigating HBV transmission events and outbreaks. Owing to these considerations there is an urgent need to develop trusted databases to store reliable and curated data on the public health aspects of HBV infections and to develop appropriate methods and tools to extract and analyse the stored data and report the information.
We present here HepSEQ (http://www.hpa-bioinfodatabases.org.uk/hepatitis_open/main.php), a freely accessible web resource on the public health aspects of HBV infection with specific focus on epidemiological, virological, clinical, nucleotide sequence and mutational aspects of HBV infection. HepSEQ is able to summarise and link large volumes of data and present those in a visually intuitive format. Moreover HepSEQ provides a resource to support detection of variants in patients from different parts of the world, to help monitor the dynamic of HBV variants during therapy and potentially to contribute to re-design of diagnostic assays.
HepSEQ
The architecture of the HepSEQ repository follows a three-tier model. A front-end Apache web server serves content to the client browsers. A middle dynamic content processing and generation layer consists of PHP, CGI and PERL scripts and specialized programs written in C (e.g. for sequence alignment). Finally a backend database has both datasets and relational database management system (RDBMS).
DATABASE
During the design and development of HepSEQ, initially all the necessary requirements for a comprehensive public health database on epidemiological, clinical and molecular markers were identified through many discussions with the stakeholders (clinicians and lab scientists) and then data modelling was undertaken. After reviewing and improving the resultant model several times, a data schema capable of catering to diverse sources and formats of data was evolved and implemented as a relational database using PostgreSQL open source database management system on a Linux operating system server. This schema consists of patient, sample, gene and mutant tables, which have one to many relationships between them. This schema enables multiple mutations to be associated with a nucleotide sequence, multiple nucleotide sequences to be associated with a sample and multiple samples to be associated with a patient.
DATA
The epidemiological, virological, clinical and nucleotide sequence data were mainly collated from the participating centres and manually checked before insertion to the database. A set of standards called ‘Caldicott Standards’ are recommended by the Department of Health, England and govern the use and transfer of patient-identifiable information from National Health Service (NHS) organizations to other NHS and non-NHS organizations (‘The Caldicott Report’ is available online at http://www.dh.gov.uk/assetRoot/04/06/84/04/04068404.pdf and ‘Confidentiality: NHS Code of Practice’ is available at http://www.dh.gov.uk/assetRoot/04/06/92/54/04069254.pdf). Any patient data that do not follow Caldicott Standards were excluded from the database. Ambiguities in data were raised with the contributors and only stored in the database once these were resolved.
WEB INTERFACE—USE AND APPLICATIONS
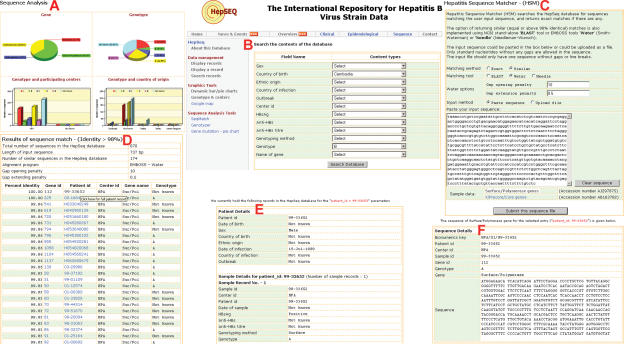
The web interface of HepSEQ has four major sections. The first of these sections contains the information pages accessed through the top navigational bar. These pages display a summary of the current contents in the repository, an overview of the HepSEQ system, and contact information. To facilitate the dissemination of news, events of interest to the public health community and latest publications on hepatitis, Really Simple Syndication (RSS) feeds from different sources are compiled and displayed on the News and Events page. Updates relating to HepSEQ are also rendered as RSS feeds and are available for download and display with a RSS newsreader from the overview page. Information pages on clinical, epidemiological and sequence display real-time data from the current database records as pie and bar charts (Figure 1).
Figure 1.
HepSEQ functionality. (A) Pie and bar charts displaying the categories of sequences in HepSEQ. (B) Search interface for HepSEQ. Sequence Matcher (C) matches a user input sequence and produces a tabular format results page (D), from which detailed individual patient record (E) or the sequence record (F) can be obtained.
The second section in HepSEQ deals with data access and submission. Users can either access all the records in a tabular form or access any detailed individual record by specifying a patient, sample, gene or mutant identifier. The search page provides all the fields available in the database and distinct entries of those. Through this page any combination of the specific fields in the database can be searched by user-created unique queries and from the resultant tabular data, individual detailed records can be viewed. Researchers interested in submitting data to HepSEQ can contact the curators by email detailing the nature, type and amount of data they wish to submit. After individually determining the best possible mechanism to submit data, the data will be bulk loaded into a temporary table and manually curated. After reconciling any inconsistencies in the data with the submitter the approved data will be migrated to the appropriate table.
The third section of the web interface provides graphical tools to dynamically generate pie and bar charts from any specified field or a pair of fields. This tool can easily find the associations between different factors (e.g. outbreak and genotype) and display those in a meaningful manner. Another tool in this section integrates the Google map API with the database and a specific parameter relating to the geographical area can be viewed.
The last section in the web interface is the sequence analytical tools. Three tools are currently available, i.e. Sequence Matcher, Genotyper and Mutation Marker.
The Sequence Matcher tool allows a user to input a DNA sequence and search it against all the sequences deposited in the HepSEQ. Protocols for identical matching as well as for matching near-identical strain sequences are available. The identical matching is implemented through the string match functions of the programming language and is very fast. Near-identical matching allows the user to pick a pairwise sequence matching algorithm from three available methods: Smith–Waterman (19), Needleman–Wunsch (20) and BLAST (21). The tabular format of the result is linked to individual records as well as alignments. The benefit of this tool is that a user-input sequence can be linked to related sequences and to potentially related cases.
The Genotyper tool assigns a genotype to a HBV sequence provided by the user. The input sequence is pairwise matched using the Needleman–Wunsch (20) algorithm against all the reference sequences and highly scoring matches (>98% sequence similarity) with statistical significance are reported. If the input sequence is not a recombinant sequence then an unambiguous genotype can be predicted to the given sequence. In this tool genotyping is done based on the sequence similarity of surface/polymerase genes of HBV. The reference sequences were assembled from the sequences downloaded from GenBank and the sequences in the HepSEQ system and validated using phylogenetic analysis. The list of reference sequences is available as Supplementary Data.
The Mutation Marker tool allows sequences grouped by different parameters (e.g. genotype) to be displayed as multiple alignments, allowing sequence differences and gaps to be visualized. This also annotates the alignment with clinically important specific mutations related to vaccine escape or antiviral resistance.
CONCLUSION AND FUTURE DIRECTIONS
The current release of HepSEQ is dedicated to be a comprehensive online resource for public health aspects of HBV infection and offers a platform for further multi-factorial analysis of HBV infection. In this current format the database system is useful as an extensive library of HBV sequences well annotated with clinical and epidemiological data.
The first priority in future HepSEQ development is to increase the number of data contributors and users and trying to reach and receive data from almost all the centres and laboratories involved in HBV infection studies. This will make HepSEQ a truly global repository of HBV infection data and a public health portal for HBV infection studies.
As the quality and consistency of the data availability is the best indicator of any database system, in future, increased focus will be towards data quality. In addition to the manual curation currently automatic scripts also report on the quality of the data. We would want to extend this to include the quality of the nucleotide sequences deposited in the system and to report on the sequencing errors that might have occurred. This has implications in assigning correct genotype to a sequence and also in the subsequent multi-factorial analysis of data. To achieve this although automated, web-based approaches can be used to a certain extent, manual curation remains the ‘gold standard’ until the acceptable parameters for automated analysis are generally agreed.
Genotype–phenotype correlations are dependant on a robust genotyping algorithm. There is a need to explore approaches other than simple percentage sequence identities between strains for genotype assignment as these can be unreliable where partial (rather than complete) genomic sequences are available. Approaches such as integrating Position Sensitive Scoring Matrices (PSSMs), recently applied to HBV (22) with the current pairwise sequence comparisons will be explored.
There is also a need to present the sequence analytical tools (genotyping and sequence alignment tools) as a webservice, so that other web-based systems could utilize these services without duplicating effort.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr Anthony Underwood for critical reading of the manuscript and Dr Manosree Chandra for the initial work on analytical tools. This project is funded by the UK Department of Health. Funding to pay the Open Access publication charges for this article was provided by Health Protection Agency, UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lee W.M. Hepatitis B virus infection: a review. N. Engl. J. Med. 1997;324:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 2.Maynard J.E. Hepatitis B: global importance and need for control. Vaccine. 1990;8(Suppl.):S18–S20. doi: 10.1016/0264-410x(90)90209-5. [DOI] [PubMed] [Google Scholar]
- 3.Mahoney F.J. Update on diagnosis, management, and prevention of hepatitis B virus infection. Clin. Microbiol. Rev. 1999;12:351–366. doi: 10.1128/cmr.12.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrari C., Missale G., Boni C., Urbani S. Immunopathogenesis of hepatitis B. J. Hepatol. 2003;39(Suppl. 1):S36–S42. doi: 10.1016/s0168-8278(03)00137-5. [DOI] [PubMed] [Google Scholar]
- 5.Kane M.A. Global status of hepatitis B immunisation. Lancet. 1996;348:696. doi: 10.1016/S0140-6736(05)65598-5. [DOI] [PubMed] [Google Scholar]
- 6.Zuckerman A.J., Zuckerman J.N. Current topics in hepatitis B. J. Infect. 2000;41:130–136. doi: 10.1053/jinf.2000.0720. [DOI] [PubMed] [Google Scholar]
- 7.Chisari F.V., Ferrari C. Hepatitis B virus immunopathogenesis. Annu. Rev. Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 8.Beasley R.P., Trepo C., Stevens C.E., Szmuness W. The e antigen and vertical transmission of hepatitis B surface antigen. Am. J. Epidemiol. 1977;105:94–98. doi: 10.1093/oxfordjournals.aje.a112370. [DOI] [PubMed] [Google Scholar]
- 9.Szmuness W., Harley E.J., Ikram H., Stevens C.E. Sociodemographic aspects of the epidemiology of hepatitis B. In: Vyas N., Cohen S.N., Schmid R., editors. Viral Hepatitis. Philadelphia: Franklin Institute Press; 1978. pp. 297–320. [Google Scholar]
- 10.Norder H., Hammas B., Lofdahl S., Courouce A.M., Magnius L.O. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J. Gen. Virol. 1992;73:1201–1208. doi: 10.1099/0022-1317-73-5-1201. [DOI] [PubMed] [Google Scholar]
- 11.Norder H., Courouce A.M., Magnius L.O. Molecular basis of hepatitis B virus serotype variations within the four major subtypes. J. Gen. Virol. 1992;73:3141–3145. doi: 10.1099/0022-1317-73-12-3141. [DOI] [PubMed] [Google Scholar]
- 12.Kidd-Ljunggren K., Miyakawa Y., Kidd A.H. Genetic variability in hepatitis B viruses. J. Gen. Virol. 2002;83:1267–1280. doi: 10.1099/0022-1317-83-6-1267. [DOI] [PubMed] [Google Scholar]
- 13.Norder H., Hammas B., Lee S.D., Bile K., Courouce A.M., Mushahwar I.K., Magnius L.O. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J. Gen. Virol. 1993;74:1341–1348. doi: 10.1099/0022-1317-74-7-1341. [DOI] [PubMed] [Google Scholar]
- 14.Norder H., Courouce A.M., Coursaget P., Echevarria J.M., Lee S.D., Mushahwar I.K., Robertson B.H., Locarnini S., Magnius L.O. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 15.Gunther S. Genetic variation in HBV infection: genotypes and mutants. J. Clin. Virol. 2006;36(Suppl.):S3–S11. doi: 10.1016/s1386-6532(06)80002-8. [DOI] [PubMed] [Google Scholar]
- 16.Francois G., Kew M., Van-Damme P., Mphahlele M.J., Meheus A. Mutant hepatitis B viruses: a matter of academic interest only or a problem with far-reaching implications? Vaccine. 2001;19:3799–3815. doi: 10.1016/s0264-410x(01)00108-6. [DOI] [PubMed] [Google Scholar]
- 17.Durantel D., Brunelle M.N., Gros E., Carrouee-Durantel S., Pichoud C., Trepo C., Zoulim F. Resistance of human hepatitis B virus to reverse transcriptase inhibitors: from genotypic to phenotypic testing. J. Clin. Virol. 2005;34(Suppl.):S34–S43. doi: 10.1016/s1386-6532(05)80008-3. [DOI] [PubMed] [Google Scholar]
- 18.Zoulim F. In vitro models for studying hepatitis B virus drug resistance. Semin. Liver Dis. 2006;26:171–180. doi: 10.1055/s-2006-939759. [DOI] [PubMed] [Google Scholar]
- 19.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 20.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Myers R., Clark C., Khan A., Kellam P., Tedder R. Genotyping Hepatitis B virus from whole- and sub-genomic fragments using position-specific scoring matrices in HBV STAR. J. Gen. Virol. 2006;87:1459–1464. doi: 10.1099/vir.0.81734-0. [DOI] [PubMed] [Google Scholar]