Abstract
SwissRegulon (http://www.swissregulon.unibas.ch) is a database containing genome-wide annotations of regulatory sites in the intergenic regions of genomes. The regulatory site annotations are produced using a number of recently developed algorithms that operate on multiple alignments of orthologous intergenic regions from related genomes in combination with, whenever available, known sites from the literature, and ChIP-on-chip binding data. Currently SwissRegulon contains annotations for yeast and 17 prokaryotic genomes. The database provides information about the sequence, location, orientation, posterior probability and, whenever available, binding factor of each annotated site. To enable easy viewing of the regulatory site annotations in the context of other features annotated on the genomes, the sites are displayed using the GBrowse genome browser interface and can be queried based on any annotated genomic feature. The database can also be queried for regulons, i.e. sites bound by a common factor.
INTRODUCTION
Regulation of the rate of transcription initiation is one of the main mechanisms through which cells regulate the expression of proteins encoded in their genomes. Transcription regulation is generally implemented through the sequence-specific binding of transcription factors (TFs) to target sites in the DNA, which are most often in the intergenic region upstream of the regulated gene.
The sequence segments recognized by TFs are generally short, i.e. typically ∼20 bp for prokaryotic TFs and ∼10 bp for eukaryotic TFs, and are normally degenerate. In spite of decades of extensive experimental work the number of experimentally known binding sites accounts only for a small fraction of the total number of functional sites that likely exist. For example, probably the most extensive data are available for Escherichia coli with ∼1000 sites that have experimental support (1), and for Sacchromyces cerevisiae with a few hundred sites that have direct experimental support (2). However, even for E.coli this constitutes probably less than one-fifth of all binding sites that exist genome wide, and only about a third of the ∼300 TFs in E.coli are represented with at least one binding site.
Computational approaches for inferring transcription factor binding sites stretch back almost two decades (3–5). However, only with the recent advent of large numbers of fully sequenced genomes, and the availability of genome-wide gene expression and chromatin immuno precipitation data has it become computationally feasible to comprehensively annotate regulatory sites genome-wide. For example, several approaches have been developed recently that identify regulatory sites by searching for significantly conserved sequence segments within multiple alignments of orthologous intergenic regions of related genomes (6–11). In this context several yeast species were sequenced recently (12,13) with the aim of identifying regulatory sites genome-wide. In addition to comparative genomic approaches large-scale ChIP-on-chip experiments have been undertaken recently in yeast to determine the intergenic regions bound by over 100 TFs (14,15). Computational approaches that combine comparative genomic analysis of orthologous intergenic regions with the analysis of these large-scale ChIP-on-chip data have led to the first comprehensive genome-wide annotations of binding sites in yeast (11,15–17).
REGULATORY SITE ANNOTATION METHODS
The methods that we use to produce the regulatory-site annotation for a given genome depend on the amounts and kinds of data that are available for that organism. For most organisms currently in SwissRegulon the only available data consist of the sequence of the genome and the genome sequences of related organisms. For these organisms our annotations are based on a careful comparison of orthologous intergenic regions from sets of related organisms as described below. For some genomes there are collections of known binding sites and we use these to build position-specific weight matrices (WMs) that represent the sequence-specificities of the TFs for which sites are available. For yeast there are also comprehensive ChIP-on-chip binding data available and we use these in combination with known sites to build a large set of WM models of yeast TFs. We use these sets of WMs to scan multiple alignments of orthologous intergenic regions genome-wide using the algorithm MotEvo (16).
IRUS: Intergenic Regions Under Selection
At the time of writing there are 354 complete microbial genomes that are available from the NCBI database (18). For all but a handful of these genomes there are no known regulatory sites, nor any ChIP-on-chip data available. However, for almost any genome in this collection one can find a number of related genomes that are close enough such that recognizable sequence homology in intergenic regions remains, even though a substantial fraction of nucleotides has been substituted since the common ancestor of the species. We have developed an automated pipeline that, starting from such a set of related genomes, predicts segments in intergenic regions that are under selection genome-wide. The details of this procedures, called IRUS, will be presented elsewhere. Here we briefly list the main steps:
We extract the genome sequences from GenBank (18) and identify orthologous genes between all pairs of species.
We reconstruct the phylogenetic tree relating the species. We first estimate the tree topology from multiple alignments of orthologous genes. Then we determine all pairwise distances from aligned third positions in 4-fold degenerate codons. Finally we fit the pairwise distances to the tree topology to obtain the branch lengths in the tree.
We construct multiple alignments of orthologous intergenic regions using T-Coffee (19).
We scan all alignments for putative regulatory sites using a probabilistic algorithm that explicitly models the evolution of regulatory sites along the phylogenetic tree. The algorithm returns posterior probabilities for each segment to contain a regulatory site and we select a set of segments with high posterior probability.
Note that the IRUS pipeline can be applied to any set of related species for which genome sequences are available.
Reconstructing WMs from known sites and ChIP-on-chip data
For E.coli and S.cerevisiae we reconstructed a set of WMs from the known binding sites in regulonDB (1) and SCPD (2) (http://egsigma.cshl.org/jian) by an automated curation procedure using the PROCSE algorithm (20). PROCSE is a probabilistic clustering algorithm that assumes the input sequences derive from an unknown number of unknown WMs and simultaneously partitions the sites into subsets that derive from a common WM, and aligns the sequences within the subsets. For each TF we also determined the site-length that maximized the overall probability of the data. For E.coli this curation lead to 97 WMs for 58 different TFs and for S.cerevisiae to 67 WMs for 62 different TFs and complexes of multiple TFs. Second, for S.cerevisiae we used the extensive binding data from (15) to infer WMs using the PhyloGibbs algorithm on alignments of orthologous intergenic regions of the Saccharomyces sensu stricto species as described in Ref. (11). Finally we combined and hand-curated the WMs resulting from the curation of the known sites and the WMs obtained with PhyloGibbs. This led to a total of 72 high confidence WMs, most of which correspond to the binding motif of a given yeast TF, whereas a small number correspond to the binding motif of a complex of yeast TFs.
MotEvo
MotEvo is a newly developed algorithm which identifies binding sites for a set of predefined WMs by scanning multiple alignments of intergenic regions (16). MotEvo exhaustively reports putative locations of binding sites and assigns a posterior probability to each reported site. For E.coli we ran MotEvo with the 97 curated WMs on multiple alignments of orthologous intergenic regions from E.coli, Salmonella typhi, Yersinia pestis KIM, Photorhabdus luminescens, and Photobacterium profundum SS9. For this dataset MotEvo reported 6237 putative sites in the E.coli genome, 1162 of which have a posterior >0.5. For S.cerevisiae we ran MotEvo with the 72 curated WMs on the multiple alignments of orthologous intergenic regions of the Saccharomyces sensu stricto species. For this dataset MotEvo reported over 85 000 putative sites, of which ∼57 000 have a posterior probability >0.1 and ∼17 000 sites having a posterior probability >0.5. For each gene MotEvo was run on the multiple alignment of intergenic regions from all species for which orthologs were available. For genes for which none of the other species have an ortholog MotEvo runs on the intergenic region of the reference species only. For these cases MotEvo effectively reduces to a WM matching algorithm.
DATABASE CONTENT
Currently the SwissRegulon database contains regulatory site annotations for the following 18 organisms: S.cerevisiae, Agrobacterium tumefaciens, Bacillus subtilis, Brucella suis, Burkholderia, Chlamydophila caviae, Corynebacterium glutamicum, Ehrlichia canis, E.coli K12, Mycobacterium tuberculosis, Neisseria meningitidis, Prochlorococcus marinus, Pseudomonas syringae, Ralstonia eutropha, Rickettsia typhi wilmington, Staphylococcus aureus, Streptococcus pneumoniae and Vibrio cholerae. Our regulatory site annotations are shown in the context of the general genome annotations provided for each of these organisms. For all organisms except for yeast the genome annotations were obtained from GenBank (18). For S.cerevisiae the genome annotation, which is significantly more extensive, was obtained from the Saccharomyces genome database (SGD) (ftp://ftp.yeastgenome.org/yeast/;21).
For all organisms except E.coli and S.cerevisiae the annotated regulatory sites are based on IRUS predictions only. For each site the genomic location, strand, sequence and the posterior probability as given by IRUS is recorded in the database. The number of regulatory sites predicted by IRUS varies from ∼750 sites for E.canis to ∼14 000 sites for Burkholderia. For E.coli and S.cerevisiae regulatory site annotations of MotEvo are given in addition to the IRUS predictions. For these regulatory sites the binding TF is also identified for each site. In addition, the database contains WM logos and regulons, i.e. lists of all annotated sites sorted by posterior probability for each TF. Finally, for S.cerevisiae the database also displays the experimentally determined binding sites from SCPD (2) and regulatory site annotations (15,17) that were downloaded from SGD. All genome-wide binding site annotations are available as flat files in gff format from the download section. For E.coli and S.cerevisiae we also provide flat files of the WMs that are used in the annotations.
DATABASE USE
The SwissRegulon database can be accessed at the address: http://www.swissregulon.unibas.ch. The database uses the Generic genome browser (GBrowse) (22) as an engine and is fully compatible with the original GBrowse. For a detailed description of GBrowse usage and features please see the original manual at the developers page (http://www.gmod.org/gbrowse). Briefly, the genome browser graphically displays a section of the genome and all features annotated on it. The user can zoom in and out and scroll through the genome and click on features to obtain more detailed information.
Users can specify a genome segment for displaying, e.g. chrII:600..1000, or query the database by entering a keyword including wild card characters, e.g. SKO*. This query will return a list of matches to the search term. For example, to find all annotated binding sites for the transcription factor RAP1 one would query the database for RAP1* (note that, beyond the binding sites this query would also return the RAP1 gene). By clicking on one of the sites in the list the user will see the section of the genome where the site occurs.
Annotated regulatory sites are displayed as rectangular boxes with an arrow inside showing the strand of the site. The posterior probability assigned to the site is represented by the intensity of the box's color. That is, the higher the posterior probability, the more intense is the color of the box. Every box is labeled by an identifier which is either the name of the TF that binds the site or a unique identifier if the site has not been assigned to any known TF.
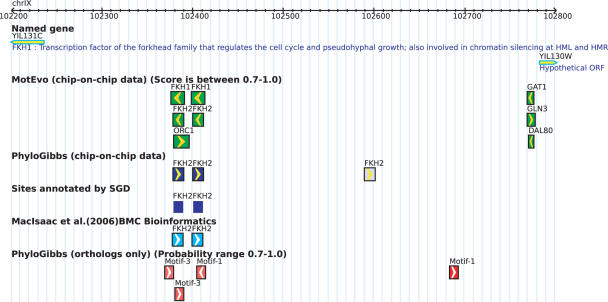
An example screen shot is shown in Figure 1. Placing the cursor on the box brings up a pop-up legend with the sequence of the site and its posterior probability. Clicking on a binding site box links to a page with detailed information about the site. For binding sites assigned to a TF this information includes the ‘regulon list’ of all sites for the same TF, and a logo of its WM. The regulon list shows for each site in the regulon the name(s) of the upstream gene(s) it regulates, the genomic coordinates of the intergenic region in which it occurs, the genomic coordinates of the site, and the posterior probability of the site. For convenient browsing the user can filter out sites according to their posterior probability. Filters are accessible under the ‘Results and Analysis’ pop-up menu.
Figure 1.
Screen shot from the SwissRegulon database for the intergenic region between the genes FKH1 and YIL130W in S.cerevisiae. Only sites with posterior probability 0.7 or larger are shown.
COMPARISON WITH EXISTING RESOURCES
There are a number of databases that collect known TF binding sites from the literature. Most of these focus on regulatory sites from a single organism, e.g. RegulonDB for E.coli (23), SCPD for S.cerevisiae (2) and the more recent regulatory site annotations based on ChIP-on-chip data (15,17), DBTBS for B.subtilis (24), AGRIS for Arabidopsis thaliana (25), and the DNase I footprint database for Drosophila melanogaster (26). Similarly, there are a number of databases that contain known binding sites in vertebrate genomes (27–29). The well-known commercial TRANSFAC database (30) probably contains the largest collection of experimentally determined binding sites from multiple organisms, mainly from eukaryotic organisms. Finally, the PRODORIC database (31) focuses on prokaryotic genomes and contains collections of known binding sites from a number of bacteria, with E.coli, B.subtilis, and Pseudomonas aeruginosa represented by a substantial number of sites. Most of these databases also contain WMs for the TFs for which known sites are available (32). Some of the databases also offer the possibility of scanning intergenic regions with these WMs, and in some cases to filter the resulting sites for conservation in related species. Additionally, databases and web servers have been made available that show the results of ‘phylogenetic footprinting’ methods (33–35), i.e. that display conservation profiles for particular sets of related genomes.
Over the last years we have developed a number of probabilistic methods (11,16,20) for rigorously combining information from known binding sites and ChIP-on-chip data with motif finding methods, and phylogenetic footprinting. By applying these methods we obtain genome-wide regulatory site annotations across different genomes using a unified methodology, which rigorously assigns quality estimates, i.e. posterior probabilities, to all predicted sites. The main aim of SwissRegulon is to make these regulatory site annotations available across as many genomes as possible, both prokaryotic and eukaryotic. In addition we make all the annotations available using a common GBrowse genome browser interface that shows the binding sites in the context of other features annotated on the genome. Through this user-friendly graphical interface the SwissRegulon resource will be useful for people researching regulatory mechanisms both experimentally and computationally.
FUTURE DEVELOPMENTS
In the near future SwissRegulon will significantly expand the number of organisms represented, especially bacterial ones. For the bacteria for which significant collections of known sites exist, e.g. B.subtilis and P.aeruginosa, we will include these into the predictions as currently done for E.coli. Eventually we also intend to include comprehensive regulatory site annotations for higher eukaryotes, i.e. vertebrate genomes, flies and worms.
Second, we are intending to incorporate ChIP-on-chip data in the SwissRegulon database in the near future. The combination between the binding site annotations and condition-specific ChIP-on-chip data will give insight into the conditions under which different sites are bound by their TFs.
Third, currently binding sites are shown on a per genome basis even though site conservation across related organisms is used in the predictions. In the future we intend to provide explicit information about conservation for each binding site and to link each binding site to the orthologous binding sites in the related genomes.
Finally, we have recently implemented a web server (http://www.phylogibbs.unibas.ch) for running the PhyloGibbs motif and regulatory site finding algorithm (11). In the future we intend to integrate these two resources. This will allow users to run PhyloGibbs on input data that was selected in the genome browser, and to see the results in the context of the existing regulatory site annotations.
Acknowledgments
The research in this study was supported by SNF grant 3152A0-105972. Funding to pay the Open Access publication charges for this article was provided by the Biozentrum, University of Basel.
Conflict of interest statement. None declared.
REFERENCES
- 1.Salgado H., Santos-Zavaleta A., Gama-Castro S., Millan-Zarate D., Blattner F., Collado-Vides J. RegulonDB (version 3.0): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 2000;28:65–7. doi: 10.1093/nar/28.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J., Zhang M.Q. SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics. 1999;15:607–611. doi: 10.1093/bioinformatics/15.7.607. [DOI] [PubMed] [Google Scholar]
- 3.Stormo G.D., Hartzell G.W. Identifying protein-binding sites from unaligned DNA fragments. Proc. Natl. Acad. Sci. USA. 1989;86:1183–1187. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey T., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 5.Lawrence C.E., Altschul S.F., Boguski M.S., Liu J.S., Neuwald A.F., Wootton J.C. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 6.McCue L.A., Thompson W., Carmack C.S., Ryan M.P., Liu J.S., Derbyshire V., Lawrence C.E. Phylogenetic footprinting of transcription factor binding sites in proteobacterial genomes. Nucleic Acids Res. 2001;29:774–782. doi: 10.1093/nar/29.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajewsky N., Socci N.D., Zapotocky M., Siggia E.D. The evolution of DNA regulatory regions for proteo-gamma bacteria by interspecies comparisons. Genome Res. 2002;12:298–308. doi: 10.1101/gr.207502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang T., Stormo G. Combining phylogenetic data with co-regulated genes to identify regulatory motifs. Bioinformatics. 2003;19:2369–2380. doi: 10.1093/bioinformatics/btg329. [DOI] [PubMed] [Google Scholar]
- 9.Moses A.M., Chiang D.Y., Eisen M.B. Phylogenetic motif detection by expectation-maximization on evolutionary mixtures. Pac. Symp. Biocomput. 2004;2004:324–335. doi: 10.1142/9789812704856_0031. [DOI] [PubMed] [Google Scholar]
- 10.Sinha S., Blanchette M., Tompa M. PhyME: A probabilistic algorithm for finding motifs in sets of orthologous sequences. BMC Bioinformatics. 2004;5:170. doi: 10.1186/1471-2105-5-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siddharthan R., Siggia E.D., van Nimwegen E. Phylogibbs: a Gibbs sampling motif finder that incorporates phylogeny. PLoS Comput. Biol. 2005;1:e67. doi: 10.1371/journal.pcbi.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 13.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 14.Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Jospeh Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., et al. Transcription regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 15.Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erb I., van Nimwegen E. IEEE Proceedings of the first International Conference on Computational Systems Biology (ICCSB), 111–118. 2006. Statistical features of yeast's transcriptional regulatory code. [Google Scholar]
- 17.Macisaac K.D., Wang T., Gordon B.D., Gifford D.K., Stormo G.D., Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Notredame C., Higgins D., Heringa J. T-Coffee: a novel method for multiple sequence alignments. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 20.van Nimwegen E., Zavolan M., Rajewsky N., Siggia E.D. Probabilistic clustering of sequences: inferring new bacterial regulons by comparative genomics. Proc. Natl Acad. Sci. USA. 2002;99:7323–7328. doi: 10.1073/pnas.112690399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong E.L., Balakrishnan R., Christie K.R., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Livstone M.S., Nash R., Park J., et al. 2006. Saccharomyces genome database. [Google Scholar]
- 22.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salgado H., Gama-Castro S., Peralta-Gil M., Diaz-Peredo E., Sanchez-Solano F., Santos-Zavaleta A., Martinez-Flores I., Jimenez-Jacinto V., Bonavides-Martinez C., Segura-Salazar J., et al. RegulonDB (version 5.0): Escherichia coli k-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makita Y., Nakao M., Ogasawara N., Nakai K. DBTBS: database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic. Acids Res. 2004;32:D75–D77. doi: 10.1093/nar/gkh074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davuluri R.V., Sun H., Palaniswamy S.K., Matthews N., Molina C., Kurtz M., Grotewold E. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergman C.M., Carlson J.W., Celniker S.E. Drosphila DNase I footprint database: a systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2004;21:1747–1749. doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed] [Google Scholar]
- 27.Blanco E., Farre D., Alba M.M., Messeguer X., Guigo R. ABS: a database of Annotated regulatory binding sites from orthologous promoters. Nucleic Acids Res. 2006;34:D63–D67. doi: 10.1093/nar/gkj116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao F., Xuan Z., Liu L., Zhang M.Q. Tred: a transcriptional regulatory element database and a platform for in silico gene regulation studies. Nucleic Acids Res. 2005;33:D103–D107. doi: 10.1093/nar/gki004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gosh D. Object-Oriented Transcription Factors Database (ooTFD) Nucleic Acids Res. 2000;28:308–310. doi: 10.1093/nar/28.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wingender E., Dietze P., Karas H., Knüppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Münch R., Hiller K., Barg H., Heldt D., Linz S., Wingender E., Jahn D. PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res. 2003;31:266–269. doi: 10.1093/nar/gkg037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vlieghe D., Sandelin A., Bleser P.J.D., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dieterich C., Wang H., Rateitschak K., Luz H., Vingron M. CORG: a database for COmparative Regulatory Genomics. Nucleic Acids Res. 2003;31:55–57. doi: 10.1093/nar/gkg007. [DOI] [PMC free article] [PubMed] [Google Scholar]