Abstract
NCBI's reference sequence (RefSeq) database (http://www.ncbi.nlm.nih.gov/RefSeq/) is a curated non-redundant collection of sequences representing genomes, transcripts and proteins. The database includes 3774 organisms spanning prokaryotes, eukaryotes and viruses, and has records for 2 879 860 proteins (RefSeq release 19). RefSeq records integrate information from multiple sources, when additional data are available from those sources and therefore represent a current description of the sequence and its features. Annotations include coding regions, conserved domains, tRNAs, sequence tagged sites (STS), variation, references, gene and protein product names, and database cross-references. Sequence is reviewed and features are added using a combined approach of collaboration and other input from the scientific community, prediction, propagation from GenBank and curation by NCBI staff. The format of all RefSeq records is validated, and an increasing number of tests are being applied to evaluate the quality of sequence and annotation, especially in the context of complete genomic sequence.
INTRODUCTION
RefSeq is a public database of nucleotide and protein sequences with feature and bibliographic annotation. The RefSeq database is built and distributed by the National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine located at the US National Institutes of Health. NCBI makes RefSeq publicly available, at no cost, over the internet via Entrez query (1) and Basic Local Alignment Search Tool (BLAST) (2,3) programs, and incorporation into a wide range of NCBI resources. The RefSeq collection is also available for FTP download as bi-monthly comprehensive releases, incremental daily updates and updates of other frequencies for select species.
NCBI builds RefSeq from the sequence data available in the archival database GenBank (4), which is a comprehensive public repository of sequences submitted to, and exchanged among, GenBank in the United States, the EMBL Data Library in the United Kingdom, and the DNA Data Bank of Japan. RefSeq records are not part of GenBank, although they can be retrieved from NCBI via the same interfaces such as Entrez Nucleotide and Entrez Protein. GenBank represents submissions from multiple groups, and thus includes sequence information generated at different times, with alternate technologies, and with diverse names and other information. In contrast, a RefSeq record is a synthesis of information and may include feature annotation, names or other data not available in the GenBank records from which it was derived. Also, RefSeq records can be updated more frequently by collaborating groups and by NCBI staff.
The RefSeq collection is unique in providing a curated, non-redundant, explicitly linked nucleotide and protein database representing significant taxonomic diversity. The collection is non-redundant in the sense that the goal is to represent distinct biological molecules that are observed for an organism, strain or haplotype. However, the molecules may themselves appear more than once in the collection if alternatively spliced transcripts encode the same protein product, if there are multiple genomic locations in one species or among species that encode the same products, or if RefSeqs are generated to represent alternate haplotypes and some mRNAs and proteins sequences are the same in all. RefSeq provides linked genomic and protein sequence records for the majority of organisms in the database; transcript records are currently limited to a subset of the eukaryotic collection. The RefSeq database provides a critical foundation for integrating sequence, genetic and functional information, and is used internationally as a standard for genome annotation. RefSeq records are accessible in several NCBI resources including Entrez Nucleotide, Protein, Gene, Map Viewer and BLAST. RefSeq records can be identified by a distinct accession format, which includes an underscore (_). A full definition of the RefSeq accession space is available on the RefSeq website (http://www.ncbi.nlm.nih.gov/RefSeq/key.html#accessions).
SCOPE
The RefSeq collection includes complete or incomplete genome sequences, transcripts and proteins. Genomic sequence records are added when whole genome submissions are submitted to GenBank and are updated as those genome sequencing projects submit updates. Genomic sequences include nuclear chromosomes, organelles, bacterial and viral genomes, and naturally occurring plasmids. RefSeq represents transcripts and proteins as represented on the GenBank submissions for many organisms; however, if whole genome sequencing projects are submitted to GenBank without annotation then NCBI may calculate annotation and provide annotated proteins in the RefSeq representation. For some eukaryotic species, RefSeq transcript and protein records are provided independently of the genome sequence and are used as a reagent to subsequently annotate the genome sequence when it becomes available. For some eukaryotic species, additional genomic sequence records are provided to represent non-transcribed pseudogenes, alternate haplotypes, gene clusters or gene-specific regions.
GROWTH
The size of the comprehensive bi-monthly RefSeq release continues to grow in pace with the large-scale genome and cDNA sequencing projects (see Table 1 and Supplementary Table 1). As of July 2006, the release included records from 3695 species and represented 2 762 164 protein records with the majority from bacterial genomes (1 990 849 proteins) and the next largest number provided for mammalian species (251 785). As new genome assemblies become available for organelles, chromosomes or complete genomes, they are incorporated into the RefSeq collection. Most organisms are represented in the collection only after some genomic sequence data (nuclear, plastid, mitochondrial or other genomic molecules) become available; however, transcript and protein records may be provided for a subset of eukaryotic organisms prior to the availability of genomic sequence data. From July 2005 (RefSeq release 12) to July 2006 (RefSeq release 18) the number of species included in the RefSeq release increased by 24%, and the total number of records increased by 46% with the largest increase, namely 62%, occurring in the protein collection.
Table 1.
Size of RefSeq release 19 per category
| Release 19, node | No. of species | Records per molecule type | ||
|---|---|---|---|---|
| Genomic | RNA | Protein | ||
| Complete | 3774 | 725 746 | 686 689 | 2 879 860 |
| Fungi | 69 | 3957 | 114 598 | 121 302 |
| Invertebrate | 231 | 212 698 | 99 939 | 101 902 |
| Microbial | 917 | 35 877 | 0 | 2 109 125 |
| Mitochondrion | 969 | 977 | 0 | 14 486 |
| Plant | 67 | 313 | 66 512 | 76 503 |
| Plasmid | 475 | 908 | 0 | 45 744 |
| Plastid | 71 | 71 | 44 | 7079 |
| Protozoa | 70 | 63 473 | 110 128 | 119 128 |
| Vertebrate_mammalian | 180 | 344 435 | 260 668 | 244 632 |
| Vertebrate_other | 459 | 62 454 | 54 092 | 60 049 |
| Viral | 1743 | 2515 | 0 | 49 598 |
ACCESS
The RefSeq collection can be accessed in multiple ways at NCBI, including by Entrez query, BLAST, FTP and links provided from NCBI databases and resources (see Supplementary Table 3). For some services, such as BLAST and queries against Entrez nucleotide and protein databases, results sets can be restricted to RefSeq records using Limits, Filters, Tabs or additional query restrictions. A subset of the available access methods is described here.
Entrez queries and links
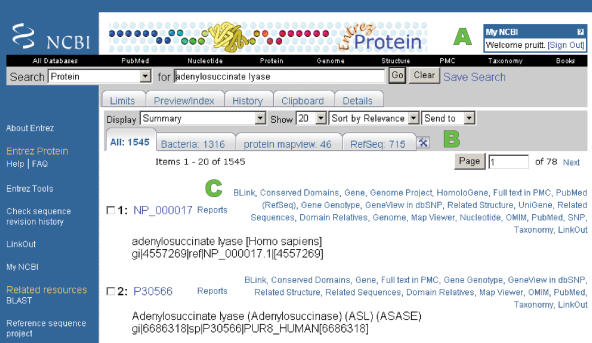
RefSeq records are included in the results returned when performing queries against the Entrez nucleotide or protein databases and the relatively new tab-oriented results page facilitates accessing the RefSeq subset (Figure 1). The display of tabs and links can be customized by logging into My NCBI. RefSeq records are extensively cross-linked with otherresources. Entrez nucleotide and protein query results include numerous links both to sets of related sequences that may include RefSeq records, and to support navigation to several additional databases and display pages (5). More links may be available from the RefSeq feature annotations as dbXrefs including links to the Consensus CDS (CCDS) project (human, mouse) and to model organisms databases such as FlyBase, MGD, WormBase or TAIR (6–9). Entrez queries can also be formatted to retrieve only RefSeq records, or to retrieve a subset of interest such as records that have been curated by either a collaborating group or by NCBI staff. For example, a query to retrieve all RefSeq nucleotide records that are annotated with a status of REVIEWED and include the name ‘BRCA1’ somewhere in the record is formatted as BRCA1 AND srcdb_refseq_reviewed[prop]. The RefSeq website provides definitions of the available property restrictions (http://www.ncbi.nlm.nih.gov/RefSeq/key.html#query).
Figure 1.
Entrez query results include records from RefSeq and GenBank (nucleotide queries) or GenPept (protein queries). (A) Users who register for MyNCBI can log on to access several services including customizing results displays. The display illustrates that user pruitt is logged in to MyNCBI. (B) Results are categorized into Tabs. The query for ‘adenylosuccinate lyase’ returns a total of 1545 records (first tab), 715 of which are RefSeq records (last tab). The display illustrates that additional tabs were added to the display to report result subsets for Bacteria and for proteins that have links to the NCBI Map Viewer. (C) Numerous links are calculated between records and can be accessed via the default ‘Links’ menu, or as shown here, the complete set of links can be shown for each record by selecting the option to display links as ‘Plain Links’ in MyNCBI. The link to ‘PubMed (RefSeq)’ returns all publications that are associated with the Entrez Gene record and thus may include a more comprehensive bibliography than that annotated on the RefSeq record.
Entrez queries from the Entrez home page, where it is possible to query against all of the Entrez databases at once, will also return results to other databases including Gene (10) and Genomes (11), which are both components of the RefSeq project. Entrez Gene integrates gene-specific annotation from RefSeq records with other sources of information, and thus provides a gene-oriented view of the genes annotated on RefSeqs. When there is sequence for a complete genome or chromosome, the data are also included in the Entrez Genome database that provides multiple tools to display and to analyze the information.
FTP
The complete RefSeq collection is made available for anonymous FTP as bi-monthly releases in conjunction with daily and cumulative updates between the release cycles. The RefSeq release is structured to provide access to the full RefSeq collection or to a portion of the collection organized by main taxonomic categories or by molecule type (e.g. mitochondrion) in order to facilitate downloads of subsets of interest. As such, the release itself is redundant as records can be found in more than one category; for example, a sequence may be included in the ‘complete’ directory and also in a taxonomic category such as the ‘plant’ directory, and optionally may occur in an organelle-oriented grouping. Extensive documentation is provided to describe the release contents including reports of files and sequences (accessions) included per category, sequences that have been removed since the previous release, species (NCBI taxonomy identifier) that have been added since the previous release, and a full description of the release structure and content. Announcements about large changes, problems and the availability of a RefSeq release are emailed to the refseq-announce email list (see Supplementary Table 2). Additional FTP data are provided for some organisms of interest, including the transcript and protein dataset for human and mouse.
COLLABORATIONS
The RefSeq project is supported by numerous collaborations that provide a variety of information including the definitions of the reference sequence standards, feature annotation and standard names. These collaborations also support the Entrez Gene database and are described in more detail in the NCBI handbook chapters for Gene, RefSeq and the Consensus CDS (CCDS) project.
Model organism databases
For some species, the RefSeq collection is curated entirely by a collaborating authoritative group that provides both the sequences and annotation. Thus RefSeq records may contain information provided by an external authoritative source and/or analyses and curation at NCBI. The collaborating group is identified on RefSeq records.
Nomenclature
Collaborations are established with official nomenclature groups when such authorities are available for an organism so that official names can be used on annotated genes. If there is no official group, data, then an effort is made to work with the research community to establish a policy for naming genes and protein products.
Consensus CDS
Annotation of genes on the human and mouse genomes is provided by multiple public resources, using different methods and resulting in information that is similar but not always identical. The human and mouse genome sequences are now sufficiently stable to start identifying those gene placements that are identical, and to make the results of those analyses public and supported as a core set by the three major public human genome browsers. The CCDS project is a collaborative effort to identify a core set of human and mouse protein coding regions that are consistently annotated and of high quality. Consistently annotated CDS regions are assigned a stable identifier and version number (e.g. CCDS1.1), which is cited on the RefSeq sequence records as a dbXref and reported in the CCDS website, Map Viewer and Entrez Gene displays (see Supplementary Table 2). The long-term goal is to support convergence toward a standard set of gene annotations on the human and mouse genomes. The CCDS set is built by consensus among the collaborating members which include (i) European Bioinformatics Institute (EBI); (ii) National Center for Biotechnology Information (NCBI); (iii) Wellcome Trust Sanger Institute (WTSI); and (iv) University of California, Santa Cruz (UCSC).
QUALITY TESTING AND CURATION
All RefSeq sequences are validated to confirm accurate nucleotide-to-protein sequence correspondence and valid ASN.1 format. Additional validation or quality testing is carried out for different subsets of the collection.
NCBI staff review and manually modify a subset of the RefSeq collection (Table 2). The goal of NCBI's manual curation is to provide accurate and full-length sequence data, to ensure accurate sequence-to-gene associations, to expand the collection by adding previously unrepresented genes and/or alternate splice products, and to provide additional feature annotation to represent mature peptide products, regions of interest, and/or to highlight less frequent biological events such as non-AUG initiation sites (12) or selenoproteins (13). The curation status is annotated on RefSeq records, as a COMMENT feature; the status terms used include model, predicted, provisional, inferred, validated and reviewed, with the latter two indicating that sequence-level curation has taken place. Curation status terms are documented on the RefSeq website (http://www.ncbi.nlm.nih.gov/RefSeq/key.html#status).
Table 2.
Number of curated protein records for select subsets
| Total | Bacteria | Plant | Viral | Coelomataa (no.) | Human | Mouse | |
|---|---|---|---|---|---|---|---|
| No. of recordsb | 2 762 164 | 1 990 849 | 72 696 | 48 799 | 78 550 | 24 874 | 19 629 |
| No. of curated | 208 783 | 120 230 | 2398 | 6472 | 20 119 | 16 049 | 2390 |
| % Curated | 7.56 | 6.04 | 3.3 | 13.26 | 25.61 | 64.52 | 12.18 |
aTranscript and protein records are curated independently of submitted annotated genomes by NCBI staff for the following organisms: Tribolium castaneum, Bombyx mori, Apis mellifera, Strongylocentrotus purpuratus, Ciona intestinalis, Danio rerio, Xenopus tropicalis, Gallus gallus, Macaca mulatta, Pan troglodytes, Homo sapiens, Canis familiaris, Felis catus, Sus scrofa, Bos Taurus, Ovis aries, Mus musculus, Rattus norvegicus, Monodelphis domestica and Takifugu rubripes.
bCuration counts per category (columns) reflect the total curation effort as contributed by either collaborating groups or NCBI staff. Curated records are annotated with a status of VALIDATED or REVIEWED.
With high-quality genomic sequence available for the human and mouse genome, review of cDNA-based RefSeqs relative to the genome has been a primary focus. The CCDS collaboration has also helped focus attention on areas where representations of mRNA and proteins sequences differ. Many tests have been added to identify possible annotation problems and thus target review to areas of most concern. QA tests include the following:
Short CDS (length < 100 amino acids).
Invalid start or stop codon.
Transcript has a stop codon in CDS.
Annotated CDS may be partial (inframe upstream start site).
Sequence is low complexity.
Protein sequence has no similarity to other protein records.
Non-consensus splice sites.
Has a very short (<5 bp) or long (>7 kb) exon, or very short (<25 bp) intron.
Single exon gene.
Gene has a spliced 5-UTR and CDS is located in the terminal exon.
Indel: transcript has insertions or deletions versus the reference genome sequence.
Mismatches: transcript has one or more mismatches versus the reference genome sequence.
Transcript does not align completely to the reference genome.
Nonsense-mediated decay (NMD) candidate (distance from stop codon to 3′-most intron following stop >55 nt).
Several of the tests were initially implemented to support the CCDS project; the scope has been expanded to include all human and mouse records. Many of the tests are designed to identify potential problems and a test failure does not necessarily indicate a real error. For example, records that do not meet minimum protein length thresholds have a higher probability of being invalid, but some very short proteins are known to exist.
Records that fail quality tests are prioritized for curation, with the highest priority given to reviewing records with potential problems in the CDS. The curation process flow includes storing database attributes to indicate that the quality test category was reviewed and the RefSeq updated, or if no problem was found with the RefSeq transcript and protein record and the reported error should be ignored, or if the problem is due to the genome assembly at that location. Assembly problems can include known gaps in the assembly and in some cases the assembled genome sequence represents a known mutation or rare polymorphism that is not the ideal sequence to represent in the transcript and protein records.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
This work was supported by the Intramural Research Program of the NIH, National Library of Medicine. Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program of the NIH, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schuler G.D., Epstein J.A., Ohkawa H., Kans J.A. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996;266:141–162. doi: 10.1016/s0076-6879(96)66012-1. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2007 doi: 10.1093/nar/28.1.15. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkl1031. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drysdale R.A., Crosby M.A., FlyBase Consortium. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blake J.A., Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E., Mouse Genome Database. Group The Mouse Genome Database (MGD): updates and enhancements. Nucleic Acids Res. 2006;34:D562–D567. doi: 10.1093/nar/gkj085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwarz E.M., Antoshechkin I., Bastiani C., Bieri T., Blasiar D., Canaran P., Chan J., Chen N., Chen W.J., Davis P., et al. WormBase: better software, richer content. Nucleic Acids Res. 2006;34:D475–D478. doi: 10.1093/nar/gkj061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkq1237. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatusova T.A., Karsch-Mizrachi I., Ostell J.A. Complete genomes in WWW Entrez: data representation and analysis. Bioinformatics. 1999;15:536–543. doi: 10.1093/bioinformatics/15.7.536. [DOI] [PubMed] [Google Scholar]
- 12.Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A.C., Vagner S. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell. 2003;95:169–178. doi: 10.1016/s0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 13.Copeland P.R. Regulation of gene expression by stop codon recoding: selenocysteine. Gene. 2003;312:17–25. doi: 10.1016/s0378-1119(03)00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]