Abstract
ECgene (http://genome.ewha.ac.kr/ECgene) was developed to provide functional annotation for alternatively spliced genes. The applications encompass the genome-based transcript modeling for alternative splicing (AS), domain analysis with Gene Ontology (GO) annotation and expression analysis based on the EST and SAGE data. We have expanded the ECgene's AS modeling and EST clustering to nine organisms for which sufficient EST data are available in the GenBank. As for the human genome, we have also introduced several new applications to analyze differential expression. ECprofiler is an ontology-based candidate gene search system that allows users to select an arbitrary combination of gene expression pattern and GO functional categories. DEGEST is a database of differentially expressed genes and isoforms based on the EST information. Importantly, gene expression is analyzed at three distinctive levels—gene, isoform and exon levels. The user interfaces for functional and expression analyses have been substantially improved. ASviewer is a dedicated java application that visualizes the transcript structure and functional features of alternatively spliced variants. The SAGE part of the expression module provides many additional features including SNP, differential expression and alternative tag positions.
INTRODUCTION
Alternative splicing (AS) is an eukaryote-specific cellular mechanism of creating diverse mRNA structures by differential use of splice sites (1). We have seen substantial progress in understanding the significance and mechanism of AS via both computational and experimental approaches. Several studies have revealed the role of AS in developmental regulation (2), evolutionary processes (3) and even in psychological behavior (4). Burge and coworkers developed computational methods to identify regulatory elements of AS—i.e. enhancers and silencers of splicing (5,6). High-throughput experimental techniques such as splice arrays have become commercially available recently.
Proper functional annotation is an essential part in understanding the role of splice variants at the genome scale (7). Many databases and applications have been developed to annotate genomes so far. European community (especially the EBI) has made significant efforts to include splice variants as a part of their Ensembl genome annotation project. Tharanaj and coworkers have developed a series of databases (ASD, AltSplice and AltTrans) by datamining GenBank sequences and PubMed literatures (8,9). AceView provides a comprehensive overview of functional and structural aspects of alternatively spliced genes for human, worm and Arabidopsis genomes (10). Lee et al. (11) developed algorithms and databases (ASAP; alternative splicing annotation project) to analyze AS at the genome-wide level. Recently they developed an algorithm to predict the full-length mRNA models which is critical in understanding the significance of a given AS at the transcript level, not at the individual exon level (12). At the time of writing, they updated the ASAP database to ASAP II which covers 17 organisms and supports comparative analysis of splice variants (http://www.bioinformatics.ucla.edu/ASAP2). Holste et al. (13) developed the Hollywood database in which the conservation of AS pattern in human and mouse can be examined. Numerous other databases (14,15) are available either to model the diverse gene structures or to predict the splice variants (e.g. see the website for the NAR database issues; http://www3.oup.co.uk/nar/database/c).
Differential expression has become an essential aspect in finding potential therapeutic targets and biomarkers. SAGE and EST data have been successfully used to find differentially expressed genes (DEG) in various organs and cancerous tissues (16,17). Lee and coworkers extended the bioinformatics search to find differentially expressed splice variants in various tissues and cancers (18,19). Recently, Gupta et al. (20) developed a database and a web server that display tissue-specific transcripts and genes using UniGene EST cluster. Such database clearly indicates the importance of understanding differential expression of alternatively spliced variants.
We developed the ECgene algorithm and the accompanying web site in 2004. The algorithm introduced a novel combination of genome-based EST clustering and graph-based transcript assembly procedures (21). The database provided functional annotations for alternatively spliced genes that included the domain, Gene Ontology (GO) and expression pattern analysis based on the EST and SAGE data (22).
In this update, we have expanded the ECgene's EST clustering and mRNA modeling to support nine organisms whose genome maps are available. The species thus included are human, mouse, rat, worm, fruit fly, zebrafish, dog, chicken and Rhesus monkey. The genome-based version provides improved EST clustering compared to the transcript-based clustering. Furthermore, mRNA modeling of splice variants is automatically incorporated in the assembly procedure. We have also developed several new applications and utilities for functional annotation of alternatively spliced genes in the human genome. Notably, a java-based viewer with several novel features visualizes AS so that users can compare splice variants efficiently. The viewer combines the advantages of the genome browser and transcript viewer in a single user interface by supporting variable intron scaling. This is in contrast to the use of two separate windows in the INTRIS program (23). Furthermore, functional domains of encoded proteins and splicing-regulatory elements are indicated in this new interface to facilitate understanding the functional significance and regulatory mechanism of AS. Expression pattern analysis includes many new features as well. We also added several new programs to identify DEG and isoforms in various organs and/or cancer tissues. Together with the new features, ECgene should represent an even more useful tool in biomarker discovery.
APPLICATIONS AND WEB INTERFACE
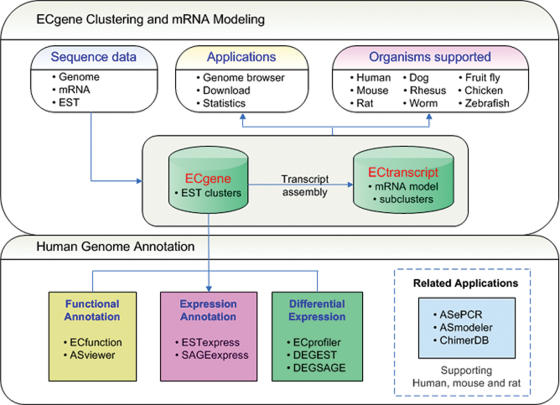
Figure 1 shows the overview of the ECgene web site. The updated version consists of two main components—expansion of ECgene clustering to various organisms and annotation of the human genome. New tools are added to examine differential expression pattern which may aid identifying tissue- and/or cancer-specific genes. Links to applications are provided inside the picture as well as in the tab menu for user convenience. Relevant databases and applications are briefly discussed below.
Figure 1.
Overview of the ECgene web site. Click on the application name launches the application.
ECgene clustering and gene modeling for alternative splicing
The ECgene algorithm was applied to nine organisms that include most of the important model organisms. This implies that we have the mRNA model and the subcluster for each splice variant, in addition to the genome-based EST clusters which are equivalent to the UniGene clusters. The result is quite similar to the TIGR Gene Indices that provides clustering and assembly for eukaryotic genomes (24). However, the genome-based method is superior to the transcript-based method in terms of clustering accuracy with a limitation that it can be applied only to organisms with the genome map. Subclusters and mRNA models are available at the ECgene download site. We also provide the ECgene genome browser that shows the genomic alignment of mRNA models and EST sequences as custom tracks in the UCSC genome browser (25). This allows users to access ample annotation tracks in the UCSC genome browser database, thereby facilitating the deduction of functional significance of each splice variant.
Table 1 compares the extent of AS for the Drosophila melanogaster genome in several databases including the FlyBase (26), DEDB (27) and ASAP II. Although the number of spliced genes is comparable between databases, ECgene shows that a significantly larger number of genes that are alternatively spliced.
Table 1.
Comparison of AS statistics for the Drosophila melanogaster genome
| DEDBa | FlyBaseb Release 4.3 | ASAP II Unigene #40 | ECgenec Part A | |
|---|---|---|---|---|
| No. of genes | — | 13 514 | 16 635 | 14 166 |
| No. of spliced genes (multi-exon genes) | 10 966 | 11 058 | 9683 | 11 657 |
| No. of transcripts | 18 567 | 19 171 | — | 26 661 |
| No. of spliced transcripts | 13 408 | 16 489 | — | 23 853 |
| No. of alternatively spliced genes | 2721 | 2814 | 1841 | 4275 |
| Percentage of alternatively spliced genes among multi-exon genes | 25 | 25 | 19 | 37 |
aCurrent version of DEDB is based on the FlyBase Release 4.2.1.
bGenes and transcripts for the FlyBase were downloaded from the UCSC table browser for the dm2 genome.
cFull statistics including ECgene part B and C is available in the website.
Functional annotation—ECfunction and ASviewer
ECfunction was developed to effectively visualize the mRNA structure and functional domains of alternatively spliced genes so that users can readily recognize any changes in the functional domains due to AS. We improved the user interface by switching to java applets that allow both zooming and intron scaling in real time. Variable intron scaling allows a seamless transition from the genome browser to the transcript or protein viewers. Thus, the detailed gene structure as well as known functional features in the genomic, mRNA and protein sequences can be readily visualized in a single user interface. Importantly, candidate splicing-regulatory signals such as the ESE (exon splicing enhancer) (5) and ESS (exon splicing silencer) (6) can be visualized with the transcript structure, which would be valuable information in studying the mechanism of AS.
ASviewer extends the features of ECfunction to support other gene models including RefSeq, Ensembl and AceView. The transcript models can be readily compared using the detailed information for exons and introns available in the baloon help. It is possible to upload the custom mRNA models and annotations into the viewer. We also provide a utility to print the genomic sequence in a similar way to the UCSC genome browser (25). The character style and color can be specified for individual mRNA models which would facilitate the detailed comparison of various predicted mRNA models.
Expression annotation—ESTexpress and SAGEexpress
ECgene's expression annotation is based on EST and SAGE data. We divided the previous version of ECexpression into two separate applications (ESTexpress and SAGEexpress) providing more specific and detailed information for each data type. ESTexpress analyzes ∼8600 human cDNA libraries and illustrates the inferred gene expression in various tissues and cancers. An option of using non-normalized libraries is also available to obtain quantitative prediction ignoring ESTs from the normalized cDNA libraries. SAGEexpress is substantially improved to provide diverse search options and detailed analysis on alternative tags. The search interface closely follows the widely used SAGEmap of NCBI and the SAGE Genie at NCI (28,29). Our tag-to-gene assignment is based on the mRNA models of ECgene. We also provide information on alternative tags stemming from alternative polyA tails, internal restriction sites and the single nucleotide polymorphisms (SNP).
Differential expression—ECprofiler, DEGEST and DEGSAGE
Special efforts have been made to facilitate the examination of the differential expression which is an issue of major importance in the field of biomarker and drug target discovery. ECprofiler is a candidate gene search system that mines EST clusters for genes with desired expression pattern and function. Specifically, the expression ontology used for cDNA library classification includes three categories—organ/tissue/cell-type, pathology and developmental stage. Both gene expression and function are implemented in ontology-based hierarchical structures. Java implementation allows users to select any combination of nodes in all categories including choice of multiple nodes and subnode expansion. We also provide a powerful search engine and diverse filtering options such as motifs, number of ESTs and libraries and the specificities.
DEGEST is a database of DEG, splice variants (isoforms) and AS events covering 52 tissues and cancer types. Chi-squared test was performed for EST clusters and subclusters from ECgene clustering to identify DEG and isoforms. DEGEST is unique in providing isoform level analysis. The background distribution of statistical test can be either the ESTs in the gene or the whole dbEST. This allows users to obtain transcripts with specific expression at the isoform level even though the gene itself has no specificity at all. DEGEST also provides specific AS events that show differential expression. AS events are classified into exon-skipping, alternative donor/acceptor sites and intron retention. Diverse filtering options are available for user convenience.
DEGSAGE tests the SAGE tags for differential expression using ∼300 SAGE libraries. We support 28 organs/tissues and cancer types. Since SAGE is inherently an mRNA-based technique, a gene may have several tags or a tag may correspond to several splice variants. We compute a representative tag to deduce expression at the gene level. The problem of tag uniqueness is included in the application.
ECprofiler and DEGSAGE run as server-client applications in real time, and the response may be slow. It is thus strongly recommended to specify the genomic region of interest within a chromosome in running ECprofiler. Although we support the genome-wide search, it should be noted that this may take over 30 min. DEGEST is a simple query system to the database that stores all results in pre-computed form for fast response.
CONCLUSION AND FUTURE DIRECTION
ECgene is an ongoing project with a collection of diverse databases and applications focused on AS. ASePCR emulates the RT–PCR experiment in various tissues. ChimerDB is a database of fusion sequences that contains chromosomal translocation. Various utilities to explore differential expression are available only for the human genome at this point. We plan to extend our functional and expression analyses to other model organisms. ECgene clustering and gene modeling will be applied to other species with a completed genome map as well. Frequent update is critical, and we plan to update ESTs on a bimonthly basis. Whole genome re-calculation takes extensive computation and will thus be updated once or twice a year depending on the amount of additional sequence data. The stable ID system is under development as well.
Acknowledgments
This work was supported by the Korean Ministry of Science and Technology through the bioinformatics research program (Grant No. 2006-01305) and by the Korean Institute for Information Technology Advancement (IITA) under the Korean Ministry of Information and Communication. Funding to pay the Open Access publication charges for this article was provided by the Korean Ministry of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Maniatis T., Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 2.Xu X., Yang D., Ding J.H., Wang W., Chu P.H., Dalton N.D., Wang H.Y., Bermingham J.R., Jr, Ye Z., Liu F., et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation–contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 3.Malko D.B., Makeev V.J., Mironov A.A., Gelfand M.S. Evolution of exon–intron structure and alternative splicing in fruit flies and malarial mosquito genomes. Genome Res. 2006;16:505–509. doi: 10.1101/gr.4236606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demir E., Dickson B.J. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121:785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 5.Fairbrother W.G., Yeo G.W., Yeh R., Goldstein P., Mawson M., Sharp P.A., Burge C.B. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeo G., Hoon S., Venkatesh B., Burge C.B. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl Acad. Sci. USA. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai M., Wang P., Boyd A.D., Kostov G., Athey B., Jones E.G., Bunney W.E., Myers R.M., Speed T.P., Akil H., et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Texier V., Riethoven J.J., Kumanduri V., Gopalakrishnan C., Lopez F., Gautheret D., Thanaraj T.A. AltTrans: transcript pattern variants annotated for both alternative splicing and alternative polyadenylation. BMC Bioinformatics. 2006;7:169. doi: 10.1186/1471-2105-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stamm S., Riethoven J.J., Le Texier V., Gopalakrishnan C., Kumanduri V., Tang Y., Barbosa-Morais N.L., Thanaraj T.A. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thierry-Mieg D., Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl. 1):11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C., Atanelov L., Modrek B., Xing Y. ASAP: the Alternative Splicing Annotation Project. Nucleic Acids Res. 2003;31:101–105. doi: 10.1093/nar/gkg029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xing Y., Yu T., Wu Y.N., Roy M., Kim J., Lee C. An expectation–maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Res. 2006;34:3150–3160. doi: 10.1093/nar/gkl396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holste D., Huo G., Tung V., Burge C.B. HOLLYWOOD: a comparative relational database of alternative splicing. Nucleic Acids Res. 2006;34:D56–D62. doi: 10.1093/nar/gkj048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leipzig J., Pevzner P., Heber S. The Alternative Splicing Gallery (ASG): bridging the gap between genome and transcriptome. Nucleic Acids Res. 2004;32:3977–3983. doi: 10.1093/nar/gkh731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollina D., Lee B.T., Tan T.W., Ranganathan S. ASGS: an alternative splicing graph web service. Nucleic Acids Res. 2006;34:W444–W447. doi: 10.1093/nar/gkl268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasmatzis G., Essand M., Brinkmann U., Lee B., Pastan I. Discovery of three genes specifically expressed in human prostate by expressed sequence tag database analysis. Proc. Natl Acad. Sci. USA. 1998;95:300–304. doi: 10.1073/pnas.95.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L., Zhou W., Velculescu V.E., Kern S.E., Hruban R.H., Hamilton S.R., Vogelstein B., Kinzler K.W. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 18.Xu Q., Lee C. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31:5635–5643. doi: 10.1093/nar/gkg786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Q., Modrek B., Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta S., Vingron M., Haas S.A. T-STAG: resource and web-interface for tissue-specific transcripts and genes. Nucleic Acids Res. 2005;33:W654–W658. doi: 10.1093/nar/gki350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim N., Shin S., Lee S. ECgene: genome-based EST clustering and gene modeling for alternative splicing. Genome Res. 2005;15:566–576. doi: 10.1101/gr.3030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim P., Kim N., Lee Y., Kim B., Shin Y., Lee S. ECgene: genome annotation for alternative splicing. Nucleic Acids Res. 2005;33:D75–D79. doi: 10.1093/nar/gki118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura K., Nishikawa T., Nagai K., Sugano S., Isogai T. Intris: a viewer for cDNA–genome alignments enabling efficient detection of splicing variants and expression profiles. Genome Inform. 2002;13:548–550. [Google Scholar]
- 24.Lee Y., Tsai J., Sunkara S., Karamycheva S., Pertea G., Sultana R., Antonescu V., Chan A., Cheung F., Quackenbush J. The TIGR gene indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res. 2005;33:D71–D74. doi: 10.1093/nar/gki064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drysdale R.A., Crosby M.A. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee B.T., Tan T.W., Ranganathan S. DEDB: a database of Drosophila melanogaster exons in splicing graph form. BMC Bioinformatics. 2004;5:189. doi: 10.1186/1471-2105-5-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lash A.E., Tolstoshev C.M., Wagner L., Schuler G.D., Strausberg R.L., Riggins G.J., Altschul S.F. SAGEmap: a public gene expression resource. Genome Res. 2000;10:1051–1060. doi: 10.1101/gr.10.7.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang P. SAGE Genie: a suite with panoramic view of gene expression. Proc. Natl Acad. Sci. USA. 2002;99:11547–11548. doi: 10.1073/pnas.192436299. [DOI] [PMC free article] [PubMed] [Google Scholar]