Abstract
Protein interaction data exists in a number of repositories. Each repository has its own data format, molecule identifier and supplementary information. Michigan Molecular Interactions (MiMI) assists scientists searching through this overwhelming amount of protein interaction data. MiMI gathers data from well-known protein interaction databases and deep-merges the information. Utilizing an identity function, molecules that may have different identifiers but represent the same real-world object are merged. Thus, MiMI allows the users to retrieve information from many different databases at once, highlighting complementary and contradictory information. To help scientists judge the usefulness of a piece of data, MiMI tracks the provenance of all data. Finally, a simple yet powerful user interface aids users in their queries, and frees them from the onerous task of knowing the data format or learning a query language. MiMI allows scientists to query all data, whether corroborative or contradictory, and specify which sources to utilize. MiMI is part of the National Center for Integrative Biomedical Informatics (NCIBI) and is publicly available at: http://mimi.ncibi.org.
1 INTRODUCTION
Both the volume and number of data sources in molecular biology are increasing rapidly. Often multiple resources provide overlapping, partial and polymorphic views of the same data. These data are stored and published in a diverse set of data sources. Each source is distinct with respect to its biological focus (e.g. SNPs, gene promoters, etc.), organism (e.g. fly) and format (e.g. tab delimited file, relational database, etc.). Even after narrowing the problem down to a subset of biological information, such as protein interaction information, there is a deluge of information. With such a rich variety of sources to choose from, a scientist who wishes to visualize the full picture concerning a particular protein must visit a myriad of sites, learn a plethora of names, aliases and identifiers, compile information from journal papers, and then piece the resulting jigsaw puzzle together. This task becomes even more onerous due to several complicating factors. First, no naming or identification scheme has been agreed upon. Thus, the scientist must painstakingly map her protein of interest to a series of different names and identifiers. Second, many interaction databases, or even lab web pages, place an interaction in the public domain even if it is supported by only one experiment. This forces scientists to search through multiple databases for conflicting or corroborating evidence. Third, heterogeneous sources storing information in their own unique formats force scientists to become programmers in order to trawl through large volumes of data and reorganize it into an understandable format. Finally, once a researcher has gathered data from several sources, sifted through it and amalgamated it, there is usually no trail left linking the data to their original sources. At this stage, if the scientist discovers mutually exclusive pieces of information existing in her amalgamated view, she has no way of making an informed decision about how to correct the data.
The work of (1,2) minimizes the burden on the user by integrating a large number of disparate sources containing information over a range of attributes, such as expression, structure and family. However, while the integration is from a large number of heterogeneous sources, it is a shallow integration. Michigan Molecular Interactions (MiMI) helps scientists search through large quantities of information by integrating all information from participating data sources through the process of deep-merging. As a result, redundant data are removed and related data are combined. Moreover, the provenance of each piece of information is tracked throughout the system, allowing scientists to choose which data to trust (3). MiMI allows users to ask more advanced questions than each of its component databases can answer independently. MiMI attempts to relieve scientists of the burden of tracking down multiple sources, mapping multiple identifiers and merging redundancies. By integrating well-known datasets, such as HPRD (4) and BIND (5), MiMI creates a deep-merged repository that is a synergy of all the merged datasets. By such integration, MiMI shows scientists when facts are corroborated by different datasets, and when facts are contradicted among datasets. Moreover, the provenance of each data item is annotated, allowing scientists to view information from only the sources they trust, and facilitating understanding of contradictory information. MiMI's integration is distinctly different from the approach used by the International Molecular Exchange Consortium (IMEx). While several of the integral components of MiMI also belong to IMEx, the tasks are very different. IMEx is attempting to increase the rate of data curation by separating curation tasks among different groups. Once curation is done, the information is shared among all. However, regardless of any cooperation between data curation sites, or partitioning of resources, there will always be some data overlap or redundancy among them. MiMI does not attempt to find new data to curate, but to augment known information by highlighting redundancy and contradictions. Additionally IMEx itself is in the very infancy of data exchange, and there is as yet no cohesive, united and deeply merged dataset produced by it.
MiMI provides a simple interface that allows new users to pose complex queries. Utilizing a simple point and click method, our user interface allows scientist to formulate advanced queries without specialized programming skills. Additionally, MiMI output complies with the PSI-MI format and Cytoscape (6), allowing users to take advantage of industry tools for viewing interactions. The following is a brief description of the underlying concepts of MiMI, as well as a detailed list of datasets employed by MiMI.
2 DATABASE CONSTRUCTION
MiMI uses XML as its data model. XML is the current lingua franca of biological data exchange, and gives the MiMI data model the flexibility to change as biological understanding increases. The physical storage of MiMI is built upon Timber (7), a native XML database. MiMI is a component of the National Center for Integrative Biomedical Informatics (http://www.ncibi.org), and is publicly available at: http://mimi.ncibi.org.
Datasets
MiMI currently has 117 549 molecules and 256 757 interactions, and is the result of integrating BIND (5), DIP (8), BioGRID (9), HPRD (4) and IntAct (10) as well as datasets from Center for Cancer Systems Biology at Harvard (11) and the Max Delbrueck Center (12). Additionally, supplementary protein information was integrated from: GO (13), InterPro (14), IPI (15), miBLAST (16), OrganelleDB (17), OrthoMCL (18) PFam (19) and ProtoNet (20).
Identity
The issue of identity is determining when two database entries refer to the same real-world object. For instance, to a human, it is obvious that an Hsp10p molecule found in yeast and listed in DIP is the same as the Hsp10p molecule found in yeast and listed in BIND. In the simplest case, identity is defined by the uniqueness of a key attribute (set); in this case, the name. However, in protein identification, no key exists across all datasets, necessitating keyless identity functions.
For example, in BioGRID, there is an entry for HSP10, and an external reference to NCBI's RefSeq NP_014663. In BIND, there is an entry for Hsp10, with an external reference to NCBI's GI 6324594. From a human perspective, it is obvious these are the same proteins with different capitalizations. However, there is no linking identifier in either BioGRID or BIND. Further searching reveals that NCBI's records hold a link between this GI and RefSeq. Given less obviously matching names for this protein, such as CPN10 (BioGRID) and Yor020p (BIND) and different external identifiers, matching identical records is not a trivial task for an individual. Combine this with the thirteen known names for this object in BIND, DIP, BioGRID and IntAct, and the human user is bound to miss incorporating some relevant data. MiMI utilizes keyless identity functions to determine which proteins represent the same real-world object. Once this identity has been determined, the entries are deep-merged.
Deep-merge
Datasets frequently have overlapping, and sometimes even contradictory information content. Our goal is to fuse information from multiple sources, even when these sources have overlapping or contradictory information, and present a cohesive result to the user. We call this process deep-merging or deep integration. (In contrast, shallow integration performs just the schema translations and groups the datasets together). To appreciate the issues involved, let us consider an example.
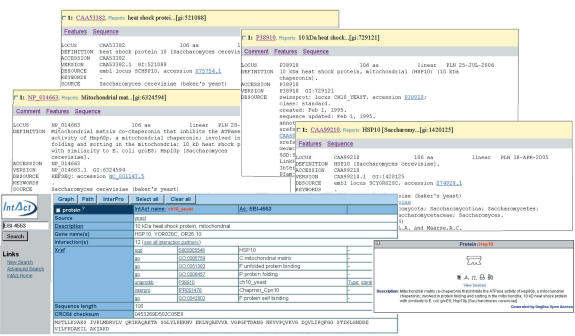
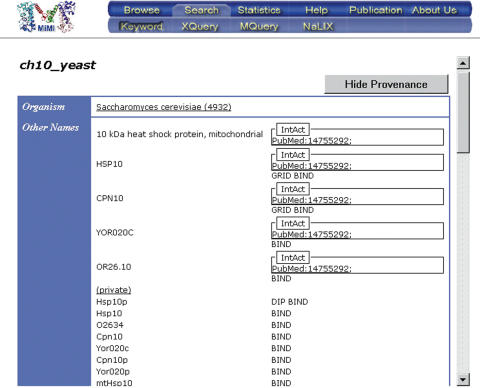
Example 1. Figure 1 shows a brief look at some of the entries for Hsp10. Each database has different identifiers and names for the molecule. Bind in 1 calls the protein Hsp10, while IntAct in 1 calls it ch10_yeast. NCBI itself has at least four versions of this protein with the exact same sequence, and different supportive information. Assuming that an appropriate identity function is found that integrates all six molecules, shallow integration would result in 15 listed interactions. However, there are only 13 non-redundant interactions reported in the datasets. A similar problem occurs for other information on the molecule, such as PTMs. Figure 2 shows a view of the resulting deep-merging process.
Figure 1.
Sample protein data for Hsp10 from IntAct, NCBI and BIND.
Figure 2.
The Hsp10 information from Figure 1 after a Deep Merge.
There is significant redundancy across data sources. Table 1 shows the number of molecules and interactions provided by each source. It also shows the resulting number of molecules and interactions after a deep-merge. For molecules there is a whopping 49% redundancy rate, while 40% of the interactions are redundant across sources.
Table 1.
Number of molecules and interactions for each source as well as total deep-merged molecules and interactions in MiMI v.2.6
| Source | # Molecules | # Interactions |
|---|---|---|
| BIND | 111 394 | 175 678 |
| IntAct | 62 667 | 67 955 |
| HPRD | 18 839 | 66 723 |
| BioGRID | 15 687 | 53 378 |
| DIP | 19 050 | 54 511 |
| Center for Cancer Systems Biology dataset | 3134 | 6726 |
| Max delbrueck center dataset | 1909 | 3269 |
| MiMI | 117 549 | 256 757 |
Provenance
Using the identity functions discussed above, datasets can be deep-merged into MiMI. However, not all datasets are created equal. Some are the result of careful curation and fact checking, while others can be from a single lab after one round of experiments. Knowing where the data came from augments its reliability. MiMI tracks the provenance of each data item, allowing the user to determine which sources to use or ignore. Moreover, database queries can use sources as a search criterion, returning only trusted information to the user.
3 DATABASE ACCESS
MiMI is stored in the Timber native XML database (7). This means that the data format follows a specific schema, and can be queried using XQuery. However, learning somebody else's internal schema is an onerous task. Additionally, writing declarative queries in a database query language, such as XQuery, has a steep learning curve. Our goal is to make MiMI easily accessible in an intuitive manner that does not rely on specialized skills.
Traditional search options
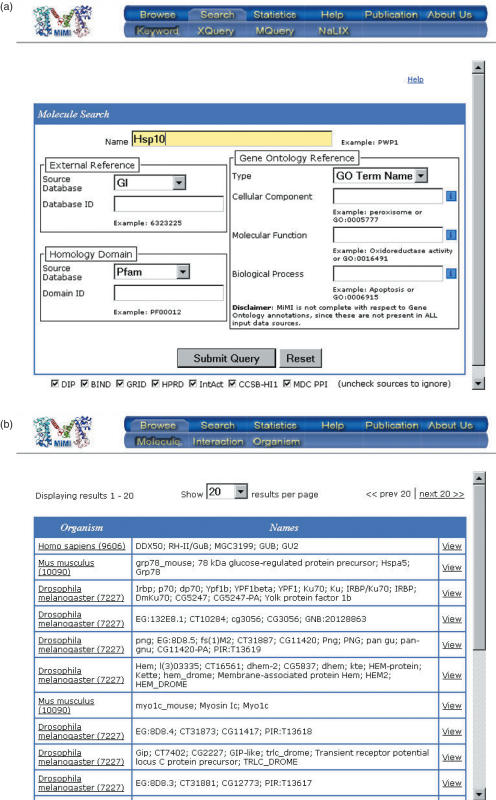
MiMI provides the user with several traditional search options, such as browse and keyword search. From the browse interface, a user may view the list of molecules, interactions or organisms included in MiMI. However, while browsing gives a general overview of the data, it is a slow way to find a particular protein. To this end, we have included a keyword query facility to allow the user quick and easy access to specific information. Figure 3a and b depict these traditional options in MiMI. This is a standard form-based search option that is similar to forms found in many online biological databases.
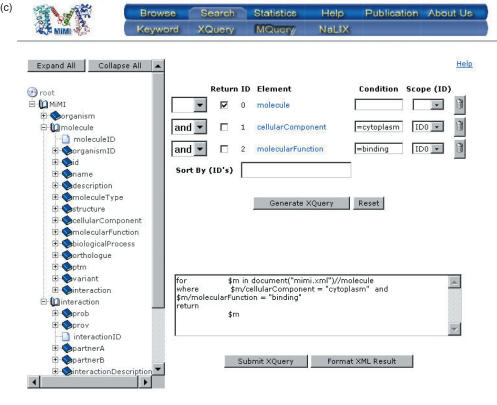
Figure 3.
Database access options: (a) Keyword query (b) browse (c) MQuery.
MQuery
Traditional approaches, such as keyword forms can be stifling and restrictive to the scientist. However, the alternative—writing a query in a declarative database query language—is prohibitive. Writing efficient XQuery is an acquired skill; browsing and keyword-based searching restricts the user's ability to pose non-trivial queries. MQuery addresses this dilemma. It combines the ease of form-based queries with the power of custom query writing (21). Additionally, the XQuery produced by MQuery is tuned to the underlying database technology and will produce an efficient XQuery.
One of the major obstacles to writing declarative queries is the need to understand the underlying document structure. MQuery allows the user to point and click on various schema elements, place conditions on them and combine these conditions conjunctively or disjunctively. Figure 3c depicts a query a beginner user could easily build by browsing through the schema on the left, clicking on fields of interest, and filling in search words. In this case, the user chose ‘cellularComponent’ and ‘moleculeFunction’, and specified that she was only interested in proteins in the cytoplasm that are involved in binding. Once the user has created a customized form according to her specifications, she presses ‘Generate XQuery’, and the appropriate XQuery statement is generated and displayed to the user. This allows the user to learn the general form of an XQuery statement, and revise the current MQuery if needed before submitting it to the database to obtain the results of the query.
Viewing information
Information from MiMI can be viewed in a variety of formats. The simplest way is to view all information via the web browser. Each page succinctly shows all recorded information for each protein as well as the provenance associated with each piece of data. A second option is to view all interactions via Cytoscape. We package all information such that it can be easily loaded and viewed in a Cytoscape browser. Finally, all information is downloadable in three different formats: XML using MiMI's internal schema, PSI-MI format (version 2.5) and plain text. Information can be downloaded from several different places: the individual molecule display page, the interaction display page and the MQuery result page.
4 FUTURE DEVELOPMENTS
MiMI is continuously changing. We are constantly looking for public datasets that would either complement the existing data or expand it. We actively encourage biologists and other users to inform us of deficiencies in either the data, or the usability of the website. Our aim is to create an essential, comprehensive and biologist-friendly database of protein interactions.
Pathway information, from sites, such as Reactome (22) will be included in the next release of MiMI. In addition to molecules and interactions, complexes, polymers, biochemical reactions and pathways will also be merged. Thus, a user viewing a complex found in both Reactome and BIND will be able to see the deep-merged record with data from both sources.
Acknowledgments
This work has been supported in part by grants R01 LM008106 and U54 DA021519 from the National Institutes of Health, by grant IIS 0219513 from the National Science Foundation, and by the Michigan Center for Biological Information.
Conflict of interest statement. None declared.
REFERENCES
- 1.Birkland A., Yona G. The BIOZON database: a hub of heterogeneous biological data. Nucleic Acids Res. 2006;34:D235–D242. doi: 10.1093/nar/gkj153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davidson S.B., Crabtree J., Brunk B.P., Schug J., Tannen V., Overton G.C., Stoeckert C.J., Jr K2/Kleisli and GUS: experiments in integrated access to genomic data sources. IBM Syst. J. 2001;40:512–531. [Google Scholar]
- 3.Buneman P., Chapman A., Cheney J. Provenance management in curated databases. ACM Sigmod. 2006:539–550. [Google Scholar]
- 4.Peri S., Navarro J.D., Amanchy R., Kristiansen T.Z., Jonnalagadda C.K., Surendranath V., Niranjan V., Muthusamy B., Gandhi T.K.B., Gronborg M., et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bader G., Betel D., Hogue C.W. BIND: the biomolecule interaction network database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jagadish H.V., Al-Khalifa S., Chapman A., Lakshmanan L.V., Nierman A., Paparizos S., Patel J.M., Srivastava D., Wiwatwattana N., Wu Y., et al. Timber: a native XML database. VLDB J. 2002;11:274–291. [Google Scholar]
- 8.Xenarios I., Salwinski L., Duan X.J., Higney P., Kin S.-M., Eisenberg D. DIP, the database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark C., Breitkreutz B.-J., Reguly T., Boucher L., Breitkreutz A., TyersStark M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia A., et al. IntAct—an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han J.D., Bertin N., Hao T., Goldberg D.S., Berriz G.F., Zhang L.V., Dupuy D., Walhout A.J., Cusick M.E., Roth F.P., et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 12.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 13.Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;8:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. Interpro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kersey P.J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. The international protein index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y.J., Boyd A., Athey B.D., Patel J.M. miBLAST: scalable evaluation of a batch of nucleotide sequence queries with blast. Nucleic Acids Res. 2005;33:4335–4344. doi: 10.1093/nar/gki739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiwatwattana N., Kumar A. Organelle DB: a cross-species database of proteinlocalization and function. Nucleic Acids Res. 2005;33:D598–D604. doi: 10.1093/nar/gki071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen F., Mackey A.J., Stoeckert C.J., Jr, Roos D.S. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. PFam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasson O., Vaaknin A., Fleischer H., Portugaly E., Bilu Y., Linial N., Linial M. ProtoNet: hierarchical classification of the protein space. Nucleic Acids Res. 2003;31:348–352. doi: 10.1093/nar/gkg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayapandian M., Jagadish H.V. In Proceedings of ICDE Conference. Atlanta, Georgia: 2006. Automating the design and construction of query forms; pp. 125–137. [Google Scholar]
- 22.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]