Abstract
Organelle DB (http://organelledb.lsi.umich.edu) is a web-accessible relational database presenting a supplemented catalog of organelle-localized proteins and major protein complexes. Since its release in 2004, Organelle DB has grown by 20% to encompass over 30 000 proteins from 138 eukaryotic organisms. Each protein in Organelle DB is presented with its subcellular localization, primary sequence and a detailed description of its function, as available. All records in Organelle DB have been annotated using controlled vocabulary from the Gene Ontology consortium. Protein localization data are inherently visual, and Organelle DB is a significant repository of biological images, housing 1500 micrographs of yeast cells carrying stained proteins. Furthermore, we report here the development of Organelle View, an extension of Organelle DB for the interactive visualization of organelles and subcellular structures in the budding yeast Saccharomyces cerevisiae. Organelle View offers a dimensional representation of a yeast cell; users can search Organelle View for proteins of interest, and the organelles housing these proteins will be highlighted in the cell image. Among other applications, Organelle View may serve as an educational aid engaging introductory biology students through a visually ‘fun’ interface. Organelle View can be accessed from the Organelle DB home page or directly at http://organelleview.lsi.umich.edu.
OVERVIEW
Since its inception in 2004, Organelle DB has provided a freely accessible information resource cataloging eukaryotic proteins that are known components of an organelle or major protein complex (1). Organelle DB presents a list of proteins organized essentially by subcellular localization and/or by organism. Each protein record housed within Organelle DB presents systematic and common gene/protein names, gene descriptions, phenotypic information (as available), biological terms from the Gene Ontology (GO) consortium, amino acid sequence and, in some cases, micrograph images (Figure 1A). To facilitate data interoperability, we have taken care to describe all protein localizations using the controlled vocabulary established by the GO consortium. In total, Organelle DB encompasses ∼60 GO localization terms; these terms have been described previously (1).
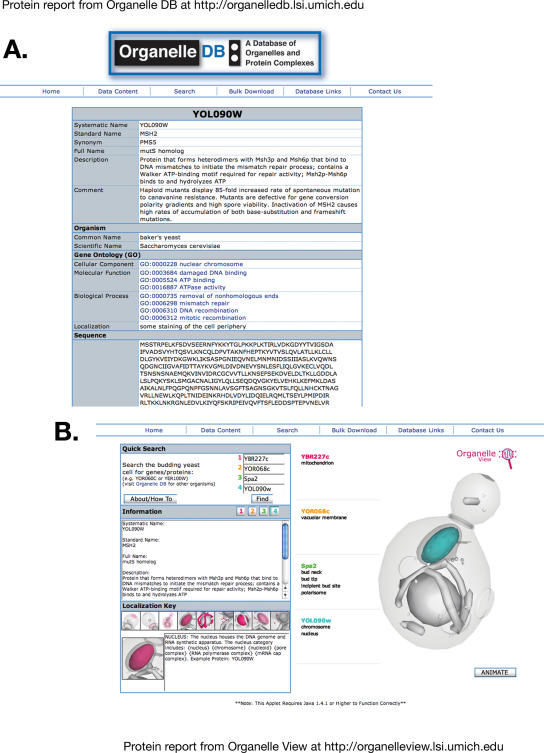
Figure 1.
Sample protein reports from Organelle DB and Organelle View. (A) Output from Organelle DB in response to a query for the yeast protein YOL090w/Msh2p, a nuclear protein involved in mismatch repair. YOL090w is the systematic name for this gene product; Msh2p is the standard or common name for this protein. (B) Output from Organelle View queried for four yeast proteins as shown. The cell image is highlighted for Msh2p/YOL090w and, thus, the nucleus appears in blue (color-coded for Msh2p).
Organelle DB has been populated in two ways. First, we have extracted protein localization data from each major model organism database [i.e. the Saccharomyces Genome Database SGD (2), the Drosophila melanogaster database FlyBase (3), the Caenorhabditis elegans database WormBase (4), the Mouse Genome Database MGD (5) and the Arabidopsis Information Resource TAIR (6)]. Localization data for human proteins and for other proteins outside of the standard model organisms have been extracted from SWISS-PROT (7) and from GO (8). Second, we have manually compiled protein localization data from large-scale and systematic studies in the budding yeast Saccharomyces cerevisiae (9–11) in supplement to localization data deposited in SGD. Since localization data have been drawn from several databases and studies, the particular source of a given protein localization record is indicated within each protein data report in the ‘Data Source’ field.
By maintaining updated localization data from these sources, we have grown Organelle DB to encompass over 31 000 proteins spanning 138 organisms across the eukaryotic kingdom. Numerical listings of protein localizations for major organisms of study are presented in Table 1. Note that we now present these data tallies by specific organism rather than by broad organismal groupings (e.g. Arabidopsis thaliana rather than ‘plants’).
Table 1.
Protein localization records in Organelle DB (as of September 2006)
| Organism | Subcellular localizations | ||||||
|---|---|---|---|---|---|---|---|
| Nucleus | Mitochondria | ER | Membrane protein | Protein complex | Miscellaneous | Total | |
| Saccharomyces cerevisiae | 2223 | 989 | 359 | 889 | 623 | 435 | 5518 |
| Arabidopsis thaliana | 1168 | 596 | 70 | 764 | 558 | 381 | 3537 |
| Drosophila melanogaster | 1383 | 498 | 101 | 863 | 554 | 260 | 3659 |
| Caenorhabditis elegans | 140 | 280 | 7 | 80 | 7 | 29 | 543 |
| Mus musculus | 1443 | 455 | 156 | 1225 | 201 | 346 | 3826 |
| Homo sapiens | 1691 | 320 | 227 | 1911 | 265 | 438 | 4852 |
| Others (132 in total) | 1847 | 8178 | 253 | 1309 | 630 | 481 | 12 698 |
| Total records | 9895 | 11 316 | 1173 | 7041 | 2838 | 2370 | 34 633 |
Note that records do not correspond exactly with proteins; one protein may have more than one record if it has been found within more than one organelle.
VISUALIZING PROTEIN LOCALIZATION DATA
Although localization data are inherently visual, text-based representations of protein localization can be difficult to understand in some cases. For example, in S.cerevisiae, a number of polarity proteins have been found localized to the bud tip; however, this descriptive term is likely unfamiliar and difficult to visualize for any researcher working with an organism other than yeast. From a simple micrograph of yeast cells, the ‘bud tip’ localization becomes clearly understood as the extreme tip of the growing bud at the opposite end of the larger, so-called mother cell. Moreover, the subtleties of a given protein localization can be lost in a simple text-based classification scheme: cytoplasmic staining can be ‘patchy’ (possibly cytoskeletal) or diffuse (from a soluble protein), with very different implications regarding protein function in each instance (9). Thus, often, the subcellular distribution of a protein is best considered by viewing an image of a cell in which the protein of interest is visualized either as a fusion to a fluorescent protein (10) or by indirect immunofluorescence staining (9). We have included this type of primary localization data in Organelle DB whenever possible; specifically, Organelle DB presents ∼1500 fluorescent micrographs of yeast cells visualized with antibodies directed against epitope-tagged proteins (indirect immunofluorescence) from our own studies of protein localization in S.cerevisiae (9,11). In addition, we welcome submissions from the scientific community of any such images for any protein reported in Organelle DB.
To further facilitate the visualization of protein localization data, we have developed an extension of Organelle DB called Organelle View. Organelle View is a scientific visualization application allowing users to dynamically generate a visual interpretation of data from Organelle DB. Organelle View presents a searchable interface with a three-dimensional representation of an archetypical cell (Figure 1B). Rather than representing organelles and subcellular structures by text, Organelle View offers an artist's rendering of a cell and its major organelles. At present, we have chosen a budding yeast cell (S.cerevisiae) as the model for Organelle View, largely because protein localization has been studied quite extensively in yeast (9,10); future versions of Organelle View will incorporate additional cell types from other organisms. Users can search Organelle View for any yeast protein, and the organelle to which that protein localizes will be highlighted in the cell image. An additional text-based summary of gene function is also presented for each searched protein. Organelle View, therefore, offers an alternative mode of presentation for the information housed in Organelle DB; it also stands as a useful educational tool, providing an easily accessible and engaging platform from which introductory biology students can explore the basics of cell biology.
DESIGN AND IMPLEMENTATION
Organelle DB was developed using the PHP server-side scripting language version 4.3.9 on a Linux server running the MySQL database version 5.0.18. We populated the most recent protein localization data from the GO database and major model organism databases [the databases described above plus the Rat Genome Database RGD (12), the Dictyostelium Database dictyBase (13) and the Zebrafish Information Network ZFIN (14)]. The scripts we implemented were configured to automatically add new genes, delete obsolete genes and update the gene information obtained from each of the source databases. We also developed a facility to add/delete/edit a particular gene per curator request. The size of our current database is 324 MB.
The Organelle View application is a web-based Java applet. This applet interfaces with the existing database Organelle DB and renders a three-dimensional model of a cell with accompanying text and dynamic functionality. The rendering code was provided by the program WireFusion (Demicron). All functionality code is written in Java and JavaScript and is provided by Nformation Design (Philadelphia, PA). All buttons and text areas outside of the applet were created using the PHP and HTML languages. Models for Organelle View were created using the open-source three-dimensional modeling program Blender. The Organelle View applet requires Java 1.4.1 to function correctly.
USING ORGANELLE DB
Organelle DB is fully searchable and presents users with a variety of options for convenient data access and retrieval. From the Organelle DB home page, users may specifically search for proteins localized to a given organelle, subcellular structure or protein complex. Additional options are provided in the Quick Search form such that users may alternatively browse records related to a single organism or gene/protein. The Quick Search form on the Organelle DB home page provides six broad protein localization groupings as follows: endoplasmic reticulum (ER), nucleus, membrane protein, mitochondrion, protein complex and others. Detailed subcategories of organelles, protein complexes and organisms may be directly accessed from our Advanced Search forms (on the ‘Search’ page at Organelle DB). These Advanced Search options offer a full list of organelles and organisms contained within Organelle DB; for example, through our Advanced Search, users may select an organelle (e.g. endoplasmic reticulum) and further select a subcategory of that organelle (e.g. integral to endoplasmic reticulum membrane). In addition, users may specify an organelle and organism, thereby limiting output to only those organelle-localized proteins from the indicated organism.
Search results are presented as a list, with protein names and a brief description of each protein indicated. By clicking on a protein name, users are taken to a full protein report (Figure 1A) containing the gene's systematic name and standard/common name, gene description including phenotypic information as available, GO classifications, amino acid sequence and any captured images supporting the reported protein localization (available for some yeast proteins in Organelle DB). We have taken particular care to maintain proper nomenclature for a given organism in presenting gene names. In cases where multiple isoforms of a given protein are reported, the amino acid sequence of each isoform is presented in Organelle DB.
As an alternative to individual search queries, users may download datasets from Organelle DB in bulk. Specifically, all data in Organelle DB may be downloaded as tab-delimited text files. In total, we offer three such files. Protein localization records from Organelle DB may be downloaded in a single file. GO annotations for each protein presented in Organelle DB are provided in a separate file. A third file provides amino acid sequences in the FASTA format for all protein entries. Multiple sequences are available for certain proteins in Organelle DB; these protein sequences can be correlated to a single protein entry in the tab-delimited text file described above through the Accession ID field.
USING ORGANELLE VIEW
Like Organelle DB, Organelle View is fully searchable. Users can enter up to four proteins in the ‘Quick Search’ form to the left of our home page (Figure 1B). The proteins with localizations will be displayed in text to the right of the search form boxes, color-coded as indicated (the first protein name and localization printed in red, etc.). The corresponding organelle for each protein will also be colored accordingly in the cell image and can be highlighted by rolling over the protein name and its localization. The cell image provided in Organelle View is an artist's rendering of a budding yeast cell; descriptive terms and organelle names related to this image are presented in the ‘Localization Key’ at the bottom left of our home page. A brief description of each cellular landmark and/or organelle is provided here; the text may be viewed by scrolling over the desired Localization Key image. The cell image can be manipulated by the cursor; e.g. the cell image can be rotated by clicking/dragging the image. Also, by clicking with the right mouse button, users can zoom in and out of the cell. Organelle View also provides much of the protein function information presented in Organelle DB. Users can view summary information regarding the function of any selected protein by clicking on the protein's corresponding number in the ‘Information’ box. The resulting text display presents systematic and standard gene/protein names, a brief functional description of the protein and any comments related to the protein's function or localization.
The color scheme described above can also be ‘animated’ if multiple proteins sharing a common localization are entered into Organelle View. By this feature, the organelle common to both proteins will shift in color in the cell image, transitioning, e.g. from red (for Protein 1) to orange (for Protein 2). This automatic color shift can be toggled on/off by clicking the ‘Animate’ button to the lower right of the cell image.
A complete tutorial describing the use of Organelle View may be accessed on-line by clicking the ‘About/How To’ button from the Organelle View home page.
APPLICATIONS AND SIGNIFICANCE
Organelle DB is a cross-species information resource for researchers utilizing nearly any eukaryotic organism of study. Data from 138 organisms are encompassed in Organelle DB. In particular, we are taking significant care to ensure that Organelle DB is fully integrated with major model organism sites and relevant external databases. Each protein report in Organelle DB is linked to the appropriate external database (i.e. the model organism sites SGD, TAIR, MGD, FlyBase, WormBase or the protein database SWISS-PROT). Thus, users can quickly drill deeper into specific proteins of interest. Furthermore, we have maintained a controlled vocabulary as much as possible in annotating Organelle DB in order to ensure significant data interoperability. We have carefully utilized proper gene/protein names for each record in Organelle DB; whenever possible, we have drawn protein names from the appropriate model organism sites in order to comply with all naming conventions for each respective organism. Users can then easily navigate between our information and relevant information in other external sites.
Collectively, the data in Organelle DB may be used by researchers in a broad swath of disciplines ranging from evolutionary biology to molecular/cellular biology and genomics/bioinformatics. Through the datasets in Organelle DB, evolutionary biologists will be able to consider organelle evolution through the eukaryota, particularly as we generate more complete datasets of eukaryotic protein localization. By integrating localization data with protein–protein interaction data, molecular, cellular and developmental biologists can ferret out higher-confidence subsets of protein interactions for further study. Researchers applying genomics and bioinformatics can use the data in Organelle DB to investigate the organelle as a functional unit, profiling and cataloging the dynamics of the organelle (i.e. its known constituent proteins) in response to cell growth and cell stress. Thus, Organelle DB is a cross-species and cross-discipline resource of general interest to the greater scientific community.
In its present form, Organelle View is a first step toward the development of non-text-based resources for the presentation of protein localization data. In addition, we view it as a particularly useful resource in the instruction of younger students (e.g. high school biology students and college undergraduates), introducing them to complicated concepts in cellular and molecular biology through an interface that is visually arresting and ‘fun’.
FUTURE DIRECTIONS
As part of our ongoing maintenance of Organelle DB, we intend to update protein localization records, by placing a particular emphasis upon proteins differentially localized during cell development and/or during cell stress responses. We also plan to modify Organelle View. We envision Organelle View as a true complement to Organelle DB—an interface for the graphical visualization of proteins within organelles and protein complexes both in a static and dynamic model of the cell. Thus, we are currently working to generate a dynamic representation of the yeast cell cycle, such that differentially localized proteins can be represented during cell cycle progression. The current prototype for Organelle View presents a budding yeast cell, but once this platform is well established, we expect to develop similar three-dimensional models for other organisms as well, providing both a broader range of cell types as well as a more detailed cell view with finer subcellular resolution.
DATABASE ACCESS
Organelle DB may be accessed freely at http://organelledb.lsi.umich.edu through the Life Sciences Institute at the University of Michigan. User support may be obtained from Organelle DB by contacting anujk@umich.edu. Please direct all technical questions and concerns to this address as well. When referencing Organelle DB and/or Organelle View, please cite this article.
Acknowledgments
We thank Hosagrahar Jagadish for his assistance in establishing this project and Harry Caul for expert data monitoring and recording. This work was supported by NSF grant DBI-0543017, American Cancer Society Research Scholar grant RSG-06-179-01-MBC and March of Dimes Basil O'Connor Starter Scholar Research Award 5-FY05-1224 (to A.K.) Funding to pay the Open Access publication charges for this article was provided by grant DBI 0543017 from the National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wiwatwattana N., Kumar A. Organelle DB: a cross-species database of protein localization and function. Nucleic Acids Res. 2005;33:D598–D604. doi: 10.1093/nar/gki071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirschman J.E., Balakrishnan R., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hong E.L., Livstone M.S., Nash R., et al. Genome Snapshot: a new resource at the Saccharomyces Genome Database (SGD) presenting an overview of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2006;34:D442–D445. doi: 10.1093/nar/gkj117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grumbling G., Strelets V. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–D488. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwarz E.M., Antoshechkin I., Bastiani C., Bieri T., Blasiar D., Canaran P., Chan J., Chen N., Chen W.J., Davis P., et al. WormBase: better software, richer content. Nucleic Acids Res. 2006;34:D475–D478. doi: 10.1093/nar/gkj061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blake J.A., Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E., Group M.G.D. The mouse genome database (MGD): updates and enhancements. Nucleic Acids Res. 2006;34:D562–D567. doi: 10.1093/nar/gkj085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhee S., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis information resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeckmann B., Bairoch A., Apweiler R., Blatter M., Estreicher A., Gasteiger E., Martin M., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar A., Agarwal S., Heyman J.A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., et al. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 11.Ross-Macdonald P., Coelho P.S., Roemer T., Agarwal S., Kumar A., Jansen R., Cheung K.H., Sheehan A., Symoniatis D., Umansky L., et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- 12.de la Cruz N., Bromberg S., Pasko D., Shimoyama M., Twigger S., Chen J., Chen C.F., Fan C., Foote C., Gopinath G.R., et al. The Rat genome database (RGD): developments towards a phenome database. Nucleic Acids Res. 2005;33:D485–D491. doi: 10.1093/nar/gki050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chisholm R.L., Gaudet P., Just E.M., Pilcher K.E., Fey P., Merchant S.N., Kibbe W.A. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res. 2006;34:D423–D427. doi: 10.1093/nar/gkj090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprague J., Bayraktaroglu L., Clements D., Conlin T., Fashena D., Frazer K., Haendel M., Howe D.G., Mani P., Ramachandran S., et al. The zebrafish information network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]