Abstract
The Human-transcriptome DataBase for Alternative Splicing (H-DBAS) is a specialized database of alternatively spliced human transcripts. In this database, each of the alternative splicing (AS) variants corresponds to a completely sequenced and carefully annotated human full-length cDNA, one of those collected for the H-Invitational human-transcriptome annotation meeting. H-DBAS contains 38 664 representative alternative splicing variants (RASVs) in 11 744 loci, in total. The data is retrievable by various features of AS, which were annotated according to manual annotations, such as by patterns of ASs, consequently invoked alternations in the encoded amino acids and affected protein motifs, GO terms, predicted subcellular localization signals and transmembrane domains. The database also records recently identified very complex patterns of AS, in which two distinct genes seemed to be bridged, nested or degenerated (multiple CDS): in all three cases, completely unrelated proteins are encoded by a single locus. By using AS Viewer, each AS event can be analyzed in the context of full-length cDNAs, enabling the user's empirical understanding of the relation between AS event and the consequent alternations in the encoded amino acid sequences together with various kinds of affected protein motifs. H-DBAS is accessible at http://jbirc.jbic.or.jp/h-dbas/.
INTRODUCTION
Alternative splicing (AS) is a phenomenon in which various combinations of exons are integrated into different types of transcripts. By utilizing AS, diverse transcripts can be produced. Although it might not be always true that all the variants are translated, this mechanism at least enables a single locus to encode functionally divergent proteins. Actual abundant cases have been reported for such diversification of the gene functions mediated by AS, in which the binding site of a growth factor receptor or an activation site of transcription factor are modified. Especially in mammals, use of AS is widespread [it is reported that 40–60% of entire human genes have AS variants (1)] and is supposed to provide a molecular basis for highly fabricated systems, such as immune systems and neural networks.
Because of the growing interests in AS, a number of databases were launched, such as ASD [http://www.ebi.ac.uk/asd/; (2)] and ASAP [http://www.bioinformatics.ucla.edu/ASAP/; (3)]. However, most of these preexisting AS databases are still incomplete in a sense that they are mainly based on the fragmented information of partially and imprecisely sequenced cDNAs (ESTs) or computationally divided information of the exons. In order to elucidate the functional relevance of the alternative variants to the protein functions, comprehensive information about the cDNA sequences is indispensable because sometimes protein motifs are embedded over a wide region of the protein sequences, and all of the combinations of the AS exons may not be allowed. Besides, for certain types of subcellular targeting signals, such as signal peptides, the position within the protein sequence is critical. Also, very recent reports, including ours (4), have demonstrated that many loci are subjected to complex patterns of AS in which two distinct genes seemed to be bridged (in which a variant uses exons from two adjacent loci), nested (in which a variant is located inside long intron of another locus) or degenerated (in which two variants use different reading frames in the shared exons. Its another name is multiple CDS): in all three cases, completely unrelated proteins are encoded by a single locus. These cases might not be regarded as alterative splicing in a strict sense. However, when those cases are also considered as extreme cases of functional diversification of a single locus and are subjected to be functional annotations, it is impossible to precisely characterize the combination of the exon usages.
Here, we introduce our new database of AS database, H-DBAS. We constructed this database exclusively using our unique dataset of completely sequenced and carefully annotated full-length cDNAs, which was produced by a human annotation meeting, H-Invitational (5,6). In H-Invitational, 56 419 cDNA sequences of human genes, which were fully sequenced with a sequence reliability higher than 99% [Phred values greater than 30; (7)] and whose potentially problematic sequences such as vectors and polyA tails were precisely trimmed, were subjected to manual annotation of AS variants. These cDNAs were clustered into 24 425 loci and of these, 6877 AS-containing loci, represented by 18 297 AS variants, were identified (4). As a specialized AS database, H-DBAS enables multifaceted analyses from various viewpoints, comprehensively aiming at elucidating functional consequences of widespread AS in human genes. [Note: We will use the word, ‘locus’, for the transcript cluster for the purpose of simplicity. However, the wording might be reconsidered, having observed highly diverse nature of the human transcriptome. Also see the reference (8)].
DATABASE CONTENTS
Data resources
In H-DBAS, the set of 167 992 so-called H-Invitational cDNAs was used (available from the URL). In addition, an option in which ASs represented by 23 210 RefSeq and 33 411 Ensembl transcripts were also considered is also implemented. In total, 167 564 transcripts were presented in the context of corresponding human genomic information as of UCSC hg17 (http://hgdownload.cse.ucsc.edu/downloads.html#human), cDNA information as of H-Invitational cDNA dataset (Table 1). The mapping and clustering procedures for the cDNAs were followed the annotation pipelines of the H-Invitational cDNAs. For details, see the help page of H-InvDB [http://jbirc.jbic.or.jp/hinv/; (9)].
Table 1.
Statistics of the data processing and of the AS variants and exons identified by genomic structure
| #Locus | #cDNA | #Total exon | #Alternative exona | #Constitutive exon | |
|---|---|---|---|---|---|
| H-Invitational cDNAs | 35 005 | 167 992 | 1 164 482b | 184 649 | 979 833 |
| Successfully mapped | 34 678 | 167 564 | 1 164 482 | 184 649 | 979 833 |
| ≥2 cDNAs per locus | 15 445 | 89 687 | 795 175 | 184 649 | 610 526 |
| Identified AS variants | 11 744 | 74 378 | 687 841 | 184 649 | 503 192 |
| Identified RASVsc | 11 744 | 38 664 | 378 024 | 98 156 | 279 868 |
| 5′-end | 7488 | 15 920 | 38 664 | 15 920 | 22 744 |
| Internal | 10 030 | 26 443 | 300 696 | 69 359 | 231 337 |
| 3′-end | 5978 | 12 877 | 38 664 | 12 877 | 25 787 |
| Retrotransposonsd | 7435 | 14 534 | 22 583 | 12 735 | 9848 |
| LINEs | 3548 | 5360 | 6620 | 3863 | 2757 |
| SINEs | 5849 | 10 188 | 14 114 | 8724 | 5390 |
| Alu elements | 4487 | 7323 | 10 240 | 6379 | 3861 |
| Identified RASVsc including full-length ORF | 11 382 | 30 389 | 311 409 | 78 078 | 233 331 |
| 5′-UTR | 6660 | 14 230 | 26 310 | 10 238 | 16 072 |
| CDS | 11 382 | 30 389 | 272 780 | 64 270 | 208 510 |
| 3′-UTR | 3519 | 5259 | 12 319 | 3570 | 8749 |
aThe number of exons was simply counted in which indicated AS relation was not associated
bUnmapped transcripts' exons could not be counted
cRepresentative AS Variants.
dThey were detected by RepeatMasker (A.F.A. Smit, R. Hubley & P. Green RepeatMasker at http://repeatmasker.org).
Data processing
Patterning alternative splicings
Using the positional information for each of the transcripts on the human genome, representative AS patterns were defined for each locus as follows. First, in order to remove possible 5′/3′-end-truncated cDNAs, we excluded cDNAs whose 5′/3′-ends were located inside the second or later exons of any other cDNAs with compatible exon structure in the same locus. We accepted the cDNAs whose 5′/3′-ends were located inside of the first/last exons and considered as variations in the exact transcriptional starting/terminating sites. We also assumed that those cDNAs whose 5′-ends were located outside of the exonic regions of any other clones could not be truncated forms of any known types of transcripts, at least [for further detailed discussion of this subject, see reference (10)]. Second, using the resulting filtered set of putative full-length cDNAs, the genomic position of each exon–intron boundary was compared with those of the other transcripts belonging to the same locus. For the comparison, a 10 bp allowance was made. If a cDNA had a part of the exonic sequence in the first/last exon inside confirmed intronic regions of the other isoforms, it was regarded as being a ‘5′/3′-end’ AS variant. If a cDNA had a part of an internal exonic sequence inside a confirmed intronic region of other isoforms, it was recognized as being an ‘internal’ AS variant (4). At this point, we removed annotated genomic rearrangement genes such as Immunoglobulin (Ig) and T-cell receptor (TCR) and anomalistic high polymorphic genes such as Major histocompatibility complex (MHC).
Merging alternative splicing patterns with functional annotations of the encoded proteins
Obtained information of patterns of AS was merged with that of detailed ORF prediction and functional annotation of H-Invitational cDNAs regarding protein motifs, GO terms, predicted subcellular localization signals and transmembrane domains. The protein motif and GO term were identified by InterProScan (11), the subcellular localization was predicted by WoLF PSORT (12) and TargetP (13) and the transmembrane domain was predicted by TMHMM (14) and SOSUI (15). For further details in functional annotation pipeline, see H-InvDB help page (http://jbirc.jbic.or.jp/hinv/). The results of the computational identification and annotation of the AS were visually inspected by the members of the AS annotation team and whenever annotations were considered to be controversial, the caveats were inserted to flag possible annotation errors.
Complex patterns of alternative splicing
Several ‘complex’ patterns of AS were defined as follows and registered in the database: (i) ‘bridged’: a locus in which two AS variants were arrayed tandemly without sharing any exons and another transcript ‘bridged’ these two isoforms, sharing at least some of its exons with both of them; (ii) ‘nested’: a locus in which CDS region of one AS variant was not shared with another variant and (iii) ‘multiple CDS’: a locus in which different ORFs >200 bp in length were annotated independently for different AS isoforms sharing at least some of the exons but not sharing any reading frame.
Current statistics
Current statistics of the database are as summarized in Tables 1 and 2 (updated from those presented in the reference (4). In total, 38 664 AS patterns were identified from 11 744 loci. When focused on the consequence of the AS to the encoded amino acid sequences, 30 389 AS variants in 11 382 loci caused changes of 97 amino acids in length on average. Further detailed statistics about how the ASs changed amino acid sequences are presented in ‘Statistics’ page in the database. Especially, 14 550 AS variants changed the protein motifs. In 14 248 cases, different GO terms were assigned to different AS variants, thus, they could be considered as good targets for further analyzing functional diversification of the genes. Similarly, AS changing subcellular localization signals and transmembrane domains were identified in 17 718 and 3995 AS variants in 5323 and 1248 loci, respectively. As for ‘complex’ AS, 2336, 3629 and 258 AS variants in 472, 1223 and 101 loci were identified and registered in the database as bridged, nested and multiple CDS, respectively.
Table 2.
Numbers of the loci in which AS variants should influence the possible protein functions
| #Locus | #cDNA | |
|---|---|---|
| AS affecting function total | 7630 | 24 092 |
| Motif-changed | 4624 | 14 550 |
| GO-changed | 4150 | 14 248 |
| Subcellular localization-changed | 5323 | 17 718 |
| Transmembrane domain-changed | 1248 | 3995 |
| Complex AS pattern total | 1512 | 5394 |
| Bridged | 472 | 2336 |
| Nested | 1223 | 3629 |
| Multiple CDS | 101 | 258 |
ACCESS TO DATABASE
Search system
A simple search form in the top page allows the user to retrieve from within H-DBAS by inputting word(s) of selected categories such as Keyword, HIX (H-Invitational cluster ID), HIT (H-Invitational transcript ID), corresponding Accession/Refseq/Emsembl ID, HUGO gene symbol and definition. In the advanced search form, the user can search the database by more detailed features of AS. The advanced search form consists of three categories: (i) ‘Genomic Location’ in which the user can specify in which chromosome and where in the chromosome the AS should be searched; (ii) ‘AS Structure’ in which the user can look for the number of representative AS variants in the locus, particular patterns of AS (such as cassette, internal acceptor, internal donor, mutually exclusive and retained intron) and their locations (5′/3′-end and internal); (iii) ‘AS Functional Annotation’ in which the user can specify the length difference of encoded protein, protein motifs, GO terms, predicted subcellular localization signals and transmembrane domains invoked by the AS: ‘Complex’ AS patterns can be also specified here. It is possible to use any combinations of the above search conditions which are within the same or different categories (Figure 1A). For example, the users can perform the search by querying the AS, which should be located on ‘chromosome 21’, having ‘internal’ ‘cassette’ exons, affecting ‘50–100 amino acids’ and ‘protein motif’. When multiple entries are hit, the user can see the Result summary and select which should be further examined (Figure 1B). Text-based summarized information can be also selected instead of showing a Java-based dynamic user interface.
Figure 1.
Search system of H-DBAS is shown. (A) Search form. H-DBAS has two sorts of search form named simple search and advanced search. The simple search allows the user to find AS locus by using keyword, H-Inv cluster ID (HIX), H-Inv transcript ID (HIT), accession number, RefSeq ID, Ensembl ID, HUGO gene symbol and definition. The advanced search allows the user to find AS locus by using various combinations of three categories such as Genomic location (i), AS structure (ii) and AS functional annotation (iii). In this search system, any combinations including simple search are available. (B) Result summary. The result of the query written in text and Figure 1A is shown.
AS Viewer
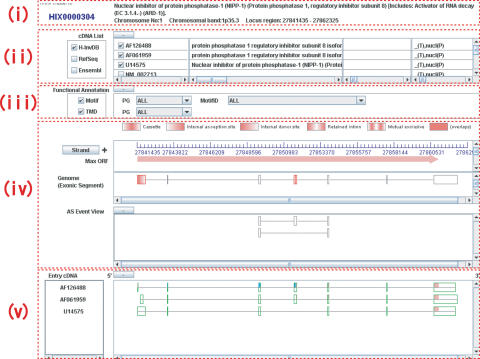
A main part of H-DBAS is a user-friendly Java-based interface, which is subjected to dynamic operations of the user (Figure 2). The browser can be zoomed from the genomic level to the sequence level (genomic/cDNA and amino acid sequences can be viewed). RefSeq and Ensembl transcripts can be viewed together with H-Invitational cDNAs as references. By using the clone view controller, the users can select which items should be viewed. Functional annotation view controllers can be used for selecting which protein motifs identified in the locus should be highlighted/erased. This page is designed so that the user can empirically recognize the positions and patterns of AS in the context of the full-length form of each transcript. It should be especially advantageous that the user can view possible influence of the AS on various kinds of protein motifs. When an AS exon in ‘Exonic Segment’ window is clicked, the positions which are regarded as mutually AS are highlighted in ‘AS Event View’ window. At the same time, if the protein motifs and transmembrane domains are identified, the corresponding exonic region of the cDNA(s) in ‘Entry cDNA’ window is colored aqua on the ORF region colored pink.
Figure 2.
AS Viewer of H-DBAS is shown. Java applet for operating AS event and checking AS exon and protein functions such as protein motif and transmembrane domain. The user can also compare with representative AS variants (RASVs) by nucleic and amino acid sequence level. AS Viewer is separated following parts: (i) Definition and genomic information of the locus; (ii) Selection function and definition of RASVs including RefSeq and Ensembl transcripts as references; (iii) Selection function of protein motif and transmembrane domain; (iv) All exons of selected RASVs are located on genome and AS exons are colored red. By clicking an AS exon in Exonic Segment field, the AS events on the location are shown in AS Event Viewer. Max ORF means total ORF range on genome of selected RASVs; (v) Selection function of AS structure on genome and on cDNA. Selected RASVs' structures are shown and these ORFs are colored pink and protein motifs and transmembrane domains are colored aqua. They are also shown nucleic and amino acid sequences by using zoom function.
Example of the search
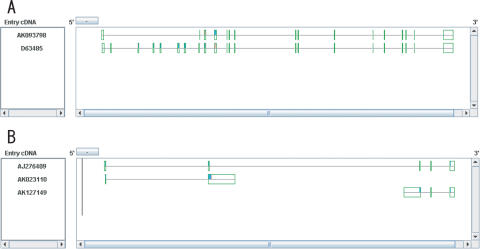
In Figure 3A, we show an example of AS affecting a motif by using AS Viewer. This is the IKK-related kinase epsilon gene. In this gene, while a cDNA (D63485) contains a prtein motif, ‘protein kinase (InterPro ID; IPR000719)’, another cDNA (AK093798) does not contain it. The lack of exons 2–7 in the latter cDNA because of cassette type AS is responsible for this putative functional difference. Figure 3B shows an example from complex AS pattern. AJ276409, which is Ssu72-like protein family protein looks as if ‘bridging’ AK127149 and AK023110 (Figure 3B), both of the latter two transcripts are of known genes and are reported to be protein-coding.
Figure 3.
Examples of the alternative splicing affecting motif (A) and bridged complex AS pattern (B) from AS Viewer in H-DBAS. Exons and introns are represented by boxes and lines. ORF region is colored pink and protein motif region is colored aqua.
Glossary and download
Use of the database as well as the archives of the raw data is freely available to anonymous public users without any restrictions. A detailed user manual and technical terms used, definitions and parameters for the annotations are precisely described in the ‘Glossary’ page in H-DBAS. The users can follow the links to further detailed information from each items displayed here. In the ‘Download’ page, archives of raw data, containing all kinds of AS information and sequence data about all AS variants in our database, are made publicly and anonymously downloadable.
FUTURE DEVELOPMENTS
We are currently interconnecting H-DBAS with H-ANGEL [http://jbirc.jbic.or.jp/hinv/h-angel/; (16)], in which gene expression patterns of the H-Invitational cDNAs are registered. We are also adding precisely annotated mouse full-length cDNA information as well and developing comparative genomics interfaces. The upcoming two major categories of extensive data will allow us to start determining how the ASs were acquired during evolution and how they fulfill the functional diversification of a single locus in various cellular circumstances. Furthermore, in the phase of further detailed experimental validation of the AS, H-DBAS should serve as an important interface for looking for cDNA clone resources, as the H-DBAS represents physical full-length cDNAs, which should serve as indispensable reagents for many kinds of experimental purposes.
Finally, we realize that we have a long way ahead for improving the web-page and database contents. We sincerely welcome any feedbacks from the users.
Acknowledgments
We thank Y. Fujii, Y. Sato, T. Habara, H. Nakaoka, F. Todokoro, Y. Imamizu, M. Ogawa and C. Yamasaki for genome mapping, ORF prediction and functional annotation of the H-Invitational cDNA dataset. We are grateful to C.Gough for critical reading of the manuscript. This research was financially supported by the Ministry of Economy, Trade and Industry of Japan (METI), the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) and the Japan Biological Informatics Consortium (JBIC). Funding to pay the Open Access publication charges for this article was provided by JBIC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 2.Stamm S., Riethoven J.J., Le Texier V., Gopalakrishnan C., Kumanduri V., Tang Y., Barbosa-Morais N.L., Thanaraj T.A. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee C., Atanelov L., Modrek B., Xing Y. ASAP: the alternative splicing annotation project. Nucleic Acids Res. 2003;31:101–105. doi: 10.1093/nar/gkg029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda J., Suzuki Y., Nakao M., Barrero R.A., Koyanagi K.O., Jin L., Motono C., Hata H., Isogai T., Nagai K., et al. Large-scale identification and characterization of alternative splicing variants of human gene transcripts using 56,419 completely sequenced and manually annotated full-length cDNAs. Nucleic Acids Res. 2006;34:3917–3928. doi: 10.1093/nar/gkl507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imanishi T., Itoh T., Suzuki Y., O'Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakao M., Barrero R.A., Mukai Y., Motono C., Suwa M., Nakai K. Large-scale analysis of human alternative protein isoforms: pattern classification and correlation with subcellular localization signals. Nucleic Acids Res. 2005;33:2355–2363. doi: 10.1093/nar/gki520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki M., Hayashizaki Y. Mouse-centric comparative transcriptomics of protein coding and non-coding RNAs. Bioessays. 2004;26:833–843. doi: 10.1002/bies.20084. [DOI] [PubMed] [Google Scholar]
- 9.Yamasaki C., Koyanagi K.O., Fujii Y., Itoh T., Barrero R., Tamura T., Yamaguchi-Kabata Y., Tanino M., Takeda J., Fukuchi S., et al. Investigation of protein functions through data-mining on integrated human transcriptome database, H-Invitational database (H-InvDB) Gene. 2005;364:99–107. doi: 10.1016/j.gene.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 10.Kimura K., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., et al. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apweiler R., Attwood T.K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton P., Park K.-J., Obayashi T., Nakai K. The 4th Annual Asia Pacific Bioinformatics Conference APBC06. 2006. Protein subcellular localization prediction with WoLF PSORT; pp. 39–48. [Google Scholar]
- 13.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 14.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 15.Hirokawa T., Boon-Chieng S., Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 16.Tanino M., Debily M.A., Tamura T., Hishiki T., Ogasawara O., Murakawa K., Kawamoto S., Itoh K., Watanabe S., de Souza S.J., et al. The human anatomic gene expression library (H-ANGEL), the H-Inv integrative display of human gene expression across disparate technologies and platforms. Nucleic Acids Res. 2005;33:D567–D572. doi: 10.1093/nar/gki104. [DOI] [PMC free article] [PubMed] [Google Scholar]