Abstract
Despite the known existence of distant-acting cis-regulatory elements in the human genome, only a small fraction of these elements has been identified and experimentally characterized in vivo. This paucity of enhancer collections with defined activities has thus hindered computational approaches for the genome-wide prediction of enhancers and their functions. To fill this void, we utilize comparative genome analysis to identify candidate enhancer elements in the human genome coupled with the experimental determination of their in vivo enhancer activity in transgenic mice [L. A. Pennacchio et al. (2006) Nature, in press]. These data are available through the VISTA Enhancer Browser (http://enhancer.lbl.gov). This growing database currently contains over 250 experimentally tested DNA fragments, of which more than 100 have been validated as tissue-specific enhancers. For each positive enhancer, we provide digital images of whole-mount embryo staining at embryonic day 11.5 and an anatomical description of the reporter gene expression pattern. Users can retrieve elements near single genes of interest, search for enhancers that target reporter gene expression to a particular tissue, or download entire collections of enhancers with a defined tissue specificity or conservation depth. These experimentally validated training sets are expected to provide a basis for a wide range of downstream computational and functional studies of enhancer function.
INTRODUCTION
The availability of the human genome sequence and large cDNA datasets, combined with efficient gene prediction methods, has led to a virtually complete list of human genes and the proteins they encode (1–4). In sharp contrast, our knowledge about the location and function of cis-regulatory elements, and in particular those of distant-acting enhancers, has been mostly confined to the results of gene-centric investigations.
The most efficient computational enhancer prediction methods available to date are based on the observation that cis-regulatory elements are often highly constrained and can therefore be identified by their conservation across evolutionary distant species. For instance, a systematic search for such conserved non-coding elements was successfully used to identify enhancers in a gene-sparse region of the human genome (5). With the availability of whole genome sequences for a growing number of vertebrates and the concurrent emergence of advanced comparative genomic tools (6,7), it is now possible to predict the location of candidate enhancers on a genome-wide scale with a specificity that makes their characterization by in vivo assays feasible. Such experimental testing requires considerable resources due to the laborious nature of the relevant functional assays. This applies especially to transgenic methods to analyze the in vivo activity of developmental enhancers. Yet, there is a pressing need for sets of enhancers with experimentally defined tissue-specific activity because these will likely be required to develop computational methods aimed at prediction of enhancer function. To be accessible and useful for such downstream applications, it is imperative that functional in vivo datasets are generated with standardized methodology and described by a consistent functional annotation. We have therefore established the VISTA Enhancer Browser as a public resource to provide access to conserved sequence elements tested for enhancer activity. The database contains results for hundreds of human candidate regions identified by comparative genomics and assayed in our own laboratory (5,8), as well as similar data from other laboratories. The purpose of this database is to provide a centralized resource for in vivo enhancer data that allows systematic mining both for gene-centric and genomic studies.
DATABASE CONTENTS
In vivo enhancer data
The core dataset of the VISTA Enhancer Browser consists of experimental in vivo data of tissue-specific enhancers. These elements are identified by their conservation between human and non-mammalian vertebrates across long (chicken and frog) or extremely long (pufferfish and zebrafish) evolutionary distances or by their unusually high conservation among mammals, such as ‘ultra’-conservation [100% identity for at least 200 bp between human, mouse and rat (9)]. The conservation properties of the currently available elements are summarized in Table 1. For more details on the experimental dataset, see Ref. 8.
Table 1.
Conservation level of 270 elements with in vivo data (as of October 2006)
| Chicken | Frog | Zebrafish | Pufferfish | Human-rodent ‘ultra’ | Datasets |
|---|---|---|---|---|---|
| + | + | + | + | + | 85 |
| + | + | + | + | 64 | |
| + | + | + | 26 | ||
| (other combinations) | 95 | ||||
The transgenic mouse assay used to determine the tissue specificity of putative enhancers has been previously described in several publications (5,8,10). Briefly, candidate elements, typically ranging in size between 200 bp and 2 kb, are amplified from human genomic DNA and cloned upstream of a heat shock protein 68 (Hsp68) promoter and a LacZ reporter gene. Importantly, the Hsp68 promoter alone lacks activity in embryonic mouse tissues (10), but drives reporter gene expression efficiently when coupled to tissue-specific enhancers. The reporter construct is linearized and injected into fertilized mouse oocytes, which are then reimplanted into pseudopregnant females. Embryos are collected on embryonic day (E) 11.5, embryo sacs are removed for genotyping and the embryos are stained for LacZ reporter gene activity (8). Note that each of these founder embryos is the result of an independent oocyte injection. Transgenic embryos can therefore be expected to carry the reporter construct stably integrated into their genome at different (random) positions. Thus, comparison of several independent transgenic embryos allows one to identify cases of expression due to positional effects (i.e. their integration near endogenous regulatory elements), which typically result in reporter activity patterns that are observed only in a single embryo out of the numerous additional transgenic animals generated in the experiment.
Elements that show consistent reporter gene expression among at least three embryos are defined as positive enhancers, whereas elements for which no reproducible pattern has been observed among a minimum of five transgenic embryos are defined as negative. It is worth noting that negatives at E11.5 do not rule out the possibility that these sequences are enhancers at other time points or physiological conditions or that their activity depends on the presence of additional cis-regulatory elements.
The tissue specificities of positive enhancers are annotated using terms that are largely consistent with the standardized nomenclature for mouse developmental anatomy (11). The reproducibility of positive enhancers is determined separately for each anatomical structure and reported in the database together with the total number of transgenic embryos obtained for each element. Thus, the reproducibility of the enhancer activity is quantified. For instance, many enhancers result in the same staining pattern in nearly all of 10 or more transgenic embryos generated, thereby indicating that they drive expression in a particular tissue very robustly and largely independent of their integration site in the genome.
Computational dataset
While the primary purpose of the VISTA Enhancer Browser is to provide a centralized resource for experimental data, it also contains a genome-wide computationally generated set of more than 145 000 conserved noncoding sequences. These elements were identified based on their highly significant (P ≤ 0.001) conservation between human, mouse and rat (6) and subsequently analyzed for conservation in chicken, frog, zebrafish and fugu to determine their conservation depth.
We plan to test subsets of these highly conserved elements in the future to further explore tradeoffs in enhancer identification success rates based on differing evolutionary conservation criteria. The major purpose of the computational dataset is, however, to provide users with lists of candidate enhancers that are easily accessible through the same interface as the experimental data and can be queried in an analogous way. The computational dataset is similar to those obtained by other comparative methods. Complementary and partially overlapping sets of highly conserved elements are, e.g. available through the VISTA Genome Browser (12), the UCSC Genome Browser (13) and the Dcode ECR Browser (14). Moreover, several web resources are available that utilize motif-based methods to predict the location of cis-regulatory elements in vertebrate genomes (15–17) and can be useful to further prioritize elements in a genome region of interest.
Data deposition by external users
The majority of experimental datasets that are currently available have been specifically generated for the VISTA Enhancer Browser (8). However, we plan to incorporate increasing amounts of published and unpublished experimental data generated by other laboratories using this lacZ-based enhancer assay in transgenic mice. These results will be available through the same interface as the in-house generated data in order to facilitate comparison of the different datasets. A software client for the upload of experimental in vivo data to this database is available from the authors upon request.
DATA RETRIEVAL
Datasets
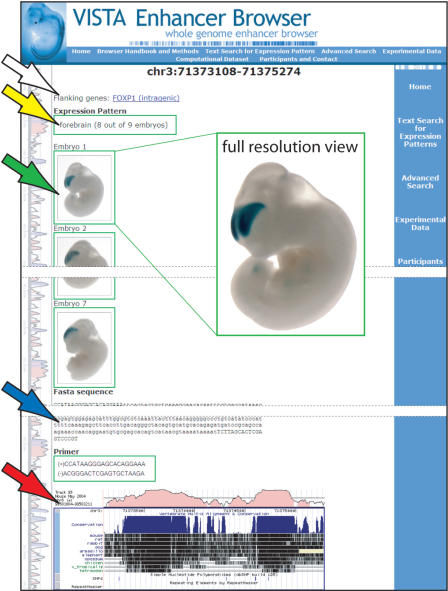
Each dataset in the VISTA Enhancer Browser consists of sequence-related information and, for experimentally tested positive elements, image data and information about enhancer tissue specificity (Figure 1).
Figure 1.
Display of experimental results for an element with reproducible enhancer activity. Flanking gene(s) are indicated at the top of the page (white arrow). The expression pattern is described by standardized anatomical terms (yellow arrow) and images of representative embryos (green arrow) are shown above the sequence of the tested human element (blue arrow) and its conservation profile (red arrow). All embryo pictures can also be viewed at high resolution (inset).
Sequence-related information includes the coordinates, the nucleotide sequence, the PCR primers used to amplify the fragment, the names of neighboring genes, and comparative information about the conservation depth of each element (Figure 1; see on-line user manual for definitions of conservation depth). For experimentally tested elements with reproducible enhancer activity, images of one or several representative embryos are provided. Embryos are shown in an overview image and, where appropriate, higher-magnification images of details of interest are provided. Moreover, the tissue specificity of reporter gene expression is presented, indicating the reproducibility separately for each anatomical structure.
Queries
Simple queries for elements in a genomic interval of interest can be initiated from the front page at http://enhancer.lbl.gov using gene symbols, accession numbers or coordinates. By default these searches are limited to elements for which experimental data are available. In the ‘Computational Dataset’ section of the database, the much larger dataset of predicted enhancers without experimental data can also be searched in a similar fashion.
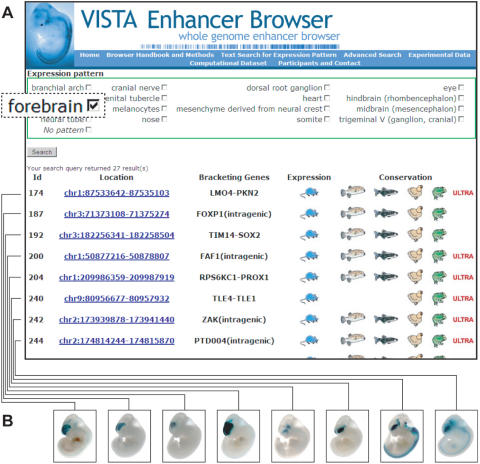
Additional search options for the experimental dataset are available in the ‘advanced query’ form. Since the expression patterns of all positive enhancers are annotated, it is possible to retrieve sets of enhancers that drive expression in certain anatomical regions or tissues (Figure 2). Checking more than one anatomical term in the query form will retrieve all elements that drive expression in any of the selected structures. In addition, the search can be limited to elements of a user-defined conservation depth. It is also possible to retrieve only elements that have no reproducible enhancer activity. When using this feature, keep in mind that the assay captures only a single developmental time-point and a negative result does not exclude the possibility that the respective element is an enhancer earlier or later in development.
Figure 2.
Retrieving enhancers that drive reporter gene expression to a user-defined anatomical structure, in this case forebrain. (A) Each row in the results table corresponds to one in vivo-assayed element. A mouse icon (blue) indicates that an element is an enhancer at E11.5. Pufferfish, zebrafish, chicken and frog icons indicate the conservation depth of the element. Images of representative embryos for each dataset (B) and more detailed information about the conservation profile of the respective element can be obtained by following the ‘location’ link.
Bulk downloads
The results of all queries are displayed in a table format that includes the coordinates and conservation depth (Figure 2). It is also possible to download all sequences of the elements retrieved by a query as a bulk sequence file in FASTA format using the ‘download’ function at the bottom of each query result. The title line for each sequence includes information if the respective element was positive or negative at E11.5.
TECHNICAL BACKGROUND
The VISTA Enhancer Browser runs on a Linux platform with mySQL 3.23 as the database engine. The web interface was developed in Perl using ImageMagick (http://www.imagemagick.org), HTML::Template (http://html-template.sourceforge.net) and overLIB (http://www.bosrup.com/web/overlib/) libraries. It runs as a CGI script under the Apache web server. All images are stored in two different resolutions (medium-sized preview and full resolution) in the file system. The web server and all parts of the database are hosted at E.O. Lawrence Berkeley National Laboratory, Berkeley, CA, USA.
AVAILABILITY
The VISTA Enhancer Browser is available to the scientific community at http://enhancer.lbl.gov. Using the web site and database is free and does not require registration. In-house generated datasets are added to the database continuously and are generally released in batches as soon as the experimental data and anatomical annotations are available. When using data retrieved from this website (e.g. sets of tissue-specific enhancers) for downstream computational applications, please cite this article or Ref. 8. If you would like to use any of the image data in publications, please contact the authors to obtain copyright.
CONCLUSIONS AND FUTURE DIRECTIONS
We have established an integrated database for functional in vivo data of human tissue-specific enhancers. The purpose of this database is to facilitate public access to a large and consistent dataset of such enhancers both for experimental and computational biologists. Candidate noncoding regions for experimental testing are identified based on their conservation between the human and other vertebrate genomes. We plan to add several hundred in-house generated datasets per year to this resource. Future goals for the VISTA Enhancer Browser include improving the interoperability with other resources and extending the tools for deposition and searching of datasets submitted by external users in order to maximize its value as a centralized resource for in vivo enhancer data.
Acknowledgments
Research was conducted at the E.O. Lawrence Berkeley National Laboratory, supported by grant HL066681 (L.A.P., I.D. and S.M.), Berkeley-PGA, under the Programs for Genomic Applications, funded by National Heart, Lung, & Blood Institute and by HG003988 (L.A.P.) funded by National Human Genome Research Institute, and performed under Department of Energy Contract DE-AC02-05CH11231, University of California. A.V. was supported by an American Heart Association postdoctoral fellowship. Funding to pay the Open Access publication charges for this article was provided by HL066681.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 4.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 5.Nobrega M.A., Ovcharenko I., Afzal V., Rubin E.M. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 6.Prabhakar S., Poulin F., Shoukry M.I., Afzal V., Rubin E.M., Couronne O., Pennacchio L.A. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M.I., Minovitsky S., Dubchak I., Holt A., Lewis K.D., et al. In vivo enhancer analysis of human conserved noncoding sequences. Nature. 2006 doi: 10.1038/nature05295. in press. [DOI] [PubMed] [Google Scholar]
- 9.Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 10.Kothary R., Clapoff S., Darling S., Perry M.D., Moran L.A., Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 11.Bard J.L., Kaufman M.H., Dubreuil C., Brune R.M., Burger A., Baldock R.A., Davidson D.R. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech. Dev. 1998;74:111–120. doi: 10.1016/s0925-4773(98)00069-0. [DOI] [PubMed] [Google Scholar]
- 12.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loots G.G., Ovcharenko I. Dcode.org anthology of comparative genomic tools. Nucleic Acids Res. 2005;33:W56–W64. doi: 10.1093/nar/gki355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanchette M., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson G., Bilenky M., Lin K., He A., Yuen W., Dagpinar M., Varhol R., Teague K., Griffith O.L., Zhang X., et al. cisRED: a database system for genome-scale computational discovery of regulatory elements. Nucleic Acids Res. 2006;34:D68–D73. doi: 10.1093/nar/gkj075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharan R., Ben-Hur A., Loots G.G., Ovcharenko I. CREME: Cis-Regulatory Module Explorer for the human genome. Nucleic Acids Res. 2004;32:W253–W256. doi: 10.1093/nar/gkh385. [DOI] [PMC free article] [PubMed] [Google Scholar]