Abstract
Publicly available database of co-expressed gene sets would be a valuable tool for a wide variety of experimental designs, including targeting of genes for functional identification or for regulatory investigation. Here, we report the construction of an Arabidopsis thaliana trans-factor and cis-element prediction database (ATTED-II) that provides co-regulated gene relationships based on co-expressed genes deduced from microarray data and the predicted cis elements. ATTED-II (http://www.atted.bio.titech.ac.jp) includes the following features: (i) lists and networks of co-expressed genes calculated from 58 publicly available experimental series, which are composed of 1388 GeneChip data in A.thaliana; (ii) prediction of cis-regulatory elements in the 200 bp region upstream of the transcription start site to predict co-regulated genes amongst the co-expressed genes; and (iii) visual representation of expression patterns for individual genes. ATTED-II can thus help researchers to clarify the function and regulation of particular genes and gene networks.
INTRODUCTION
High-throughput techniques have produced vast amounts of sequence, expression and structure data. One of these techniques, DNA microarray analysis, produces information on relative expression levels for thousands of genes simultaneously. In addition, large collections of microarray data contain information about concerted changes in transcript levels in these datasets beyond the original purpose of each dataset. Microarray data have been collected in several general repositories, including ArrayExpress (1), Gene Expression Omnibus (GEO) (2) and the Center for Information Biology Gene Expression Database (CIBEX) (3). However, it is generally difficult for experimental researchers who lack bioinformatics expertise to retrieve the information they seek.
For the model plant Arabidopsis thaliana, The Arabidopsis Information Resource (TAIR) (4) and the Nottingham Arabidopsis Stock Centre Arrays (NASCArrays) (5) are species-specific repositories for microarray data. Although it is easy to search the data for individual genes or samples, it is still difficult to retrieve gene-to-gene relationships or simply browse gene expression patterns. Several databases have become available as secondary database for microarray data. The comprehensive systems-biology database (CSB.DB) (6), Botany Array Resource (BAR) (7), Arabidopsis Co-expression Tool (ACT) (8) and Genevestigator (9) provide co-expressed gene relationships calculated from the array data stored in TAIR and/or the NASCArrays. Such gene relationship information is valuable for predicting gene function, because co-expressed genes are often involved in the same or related pathways. Approaches for specifying experimental targets using such co-expressed relationships have already been reported (10–13). Each database has its own means of displaying co-expression data. One of the tools provided in ACT, ‘clique finder’ returns a group of co-expressed genes from the co-expressed gene network, although it does not show the network itself (8). Visualized expression patterns are provided in BAR (7), because a gene expression pattern is difficult to comprehend from lists of expression values. Computational prediction of cis elements is another important feature for microarray analysis. Interactive cis-element prediction from co-expressed genes is also provided in BAR (7). It could enforce the estimation of regulatory scheme of the target genes deduced from the limited amount of experimentally determined cis elements, which are stored in cis-element databases (14,15).
Although these databases are well equipped with such user-friendly tools, additional interfaces are still required. The following three points need to be addressed: (i) Co-expressed gene lists are too long to allow visualization of complete co-expression patterns. (ii) Although the color-coded ‘heat maps’ provided by BAR (7) are suitable for comparison of a set of genes, they do not represent quantitative expression changes well. (iii) The interactive cis-element prediction tool provided by BAR (7) can extract cis elements from the user-selected genes, but the results are hard to exhaustively compare between different queries.
To support the needs of non-bioinformatics experts, we constructed a database named A.thaliana trans-factor and cis-element prediction database (ATTED-II) for retrieving gene-to-gene relationships similar to the other databases for co-expressed genes. ATTED-II contains the following three original aspects. (i) Network representation of co-expressed gene relationships, which, in addition to the original lists of co-expressed genes, facilitates understanding of the structural basis of gene co-expression (16). (ii) Stored pre-calculated results for cis-element prediction are linked to every gene and every functional category, and are presented along with several characteristics of the cis elements. (iii) The gene expression patterns are graphically represented—the display of bar graphs for individual genes makes it easy to see quantitative expression changes for many experimental conditions.
ATTED-II is based on the framework of the A.thaliana tissue-specific expression database (ATTED), which was opened in 2003 as the repository for our original data for tissue-specific gene expression (17). With the subsequent availability of public microarray data, we imported these data into the original ATTED and stored the calculated co-expression information and the predicted cis elements. ATTED-II now contains co-expressed gene networks for 22 263 loci and for 1102 functional categories as well as predicted cis elements represented by 304 heptamers. All expression data for these contents are GeneChip data (25k) released by TAIR (4), update of which will be incorporated into ATTED-II. In the following sections, the contents of ATTED-II and its usefulness are described with examples for obtaining information on co-expressed genes (Figure 1) and predicted cis elements (Figure 2).
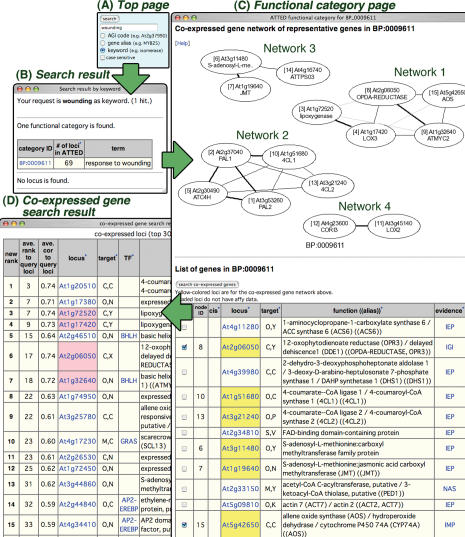
Figure 1.
Screenshots of ATTED-II pages relevant to the search for wounding-responsive gene groups. (A) Top page. (B) Search results. (C) Functional category page. Four co-expressed gene groups were found for wounding response. (D) Results of the co-expressed gene search. ‘Target’ in tables in (C) or (D) indicates predicted subcellular localization. For example, ‘C,C’ indicates both TargetP and WoLF PSORT predict that the gene product is localized in chloroplasts. Yellow-colored AGI codes in the table in (C) indicate genes in the co-expressed gene network. Pink-colored AGI codes in the table in (D) indicate query genes used to search for co-expressed genes.
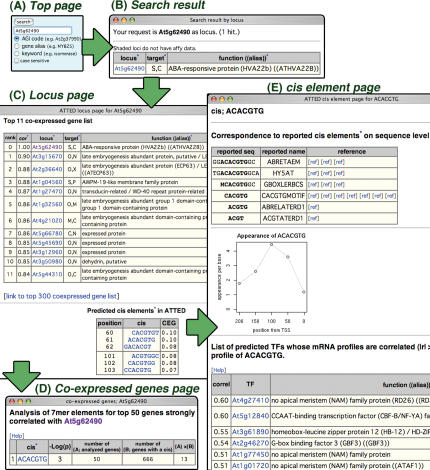
Figure 2.
Screenshots of ATTED-II pages relevant to the search for a predicted regulatory scheme of At5g62490. (A) Top page. (B) Search results. (C) Locus page. The co-expressed gene list and predicted cis elements for At5g62490. (D) Co-expressed genes page. Results of a statistical test for the presence of a cis element in the 50 co-expressed genes. (E) Cis elements page and detailed information for ACACGTG. For simplicity, some graphs are omitted in (C) and (E).
DATABASE CONTENTS
ATTED-II contains four types of pages; locus page, co-expressed genes page, cis element page and functional category page.
The locus page contains information for each of 31 128 loci, which covers all Arabidopsis protein-coding loci defined by TAIR (4). Of these loci, 22 263 have gene expression patterns measured by GeneChip. For each locus, co-expressed genes are calculated and the 12 most strongly correlated genes are represented in the locus page as a network and a list. These co-expressed genes are supported by the following information provided in the same locus page: organized functional gene annotation from other databases, subcellular localization predicted by TargetP (18) and WoLF PSORT (19), and graphs of the gene expression patterns. Predicted cis elements in the 200 bp region upstream from the transcription start site (TSS) are also shown to support co-expressed gene relationships (Figure 2C). The upstream sequence used in ATTED-II is obtained from TAIR (4), where for the loci without TSS annotation the sequences from translational start site are provided. Details about the predicted cis elements can be obtained in the cis element page.
A co-expressed genes page is prepared for each of the 22 263 loci for which there are gene expression data. This page contains a longer list of co-expressed genes (300 genes) than that found in locus page and presents statistical test results of the nearest 50 genes for functional categories and for cis-element appearances (Figure 2D). For extended analysis, whole lists—including more than 300 co-expressed genes—are downloadable from ATTED-II as tab-delimited text files.
The cis element page provides information for each predicted cis element. In this prediction, cis elements are represented by heptamers. The extracted heptamers are matched with reported cis elements stored in the Database of Plant Cis-acting Regulatory DNA Elements (PLACE) (14) (Figure 2E). The cis-element profiles for position and for experimental conditions are provided to reduce false positives and to consider biological functions (Figure 2E). Also provided are results of a statistical test for functions of the genes having a predicted cis element and a list of transcription factors predicted to bind the cis element. List of transcription factors in ATTED-II were downloaded from AGRIS (20).
A functional category page is constructed for each gene function category using the 3481 terms defined by GO (21), KEGG (22), AraCyc (23) and KaPPA-View (24). The 15 most significant co-expressed genes are selected from the genes annotated by each functional term, and co-expressed gene networks are drawn (Figure 1C). Because two-thirds of the categories have no significantly co-expressed genes, the networks are drawn for 1102 categories in total. Lists of all genes in the categories are described under the network (Figure 1C). The method for selecting the genes used to draw the networks is described in the help page in ATTED-II.
EXAMPLE 1: OBTAINING CO-EXPRESSED GENES FOR A PARTICULAR FUNCTION
As an example, consider the study of plant defense systems for wounding and injury. As a non-motile organism, a plant cannot move to a more favorable place but must bear and adapt to its environments using complex defence mechanisms. Identification of the genes in a system is the first step for deeper understandings of the system. For gene identification, it is efficient to list and classify genes of co-functional candidates using co-expressed genes (10–13). For this purpose, ATTED-II could be used as follows (Figure 1). (Note that larger figures for this example are shown in the help page in ATTED-II.).
Search ‘wounding’ as a keyword using the search form to the right of the title logo in ATTED-II (Figure 1A).
As a result, a GO category ‘response to wounding’ is found. Click the ‘Category ID’ (Figure 1B).
- The functional category page for ‘response to wounding’ provides co-expressed gene networks of 15 representative genes in this category (Figure 1C). Each circle represents a co-expressed gene. The AGI code, which is locus ID in Arabidopsis (25), a short description and the node ID are enclosed by the circle. The thickness of the lines connecting the circles corresponds to the relative strength of co-expression. The table under the network contains longer descriptions, where the genes in the networks are numbered by node IDs and highlighted by yellow-colored AGI codes. Four networks of co-expressed genes are drawn for this category. To consult each network, the information of the external links in each locus page is useful in addition to the information presented in ATTED-II. In this example, publications for each locus provided by TAIR (4) were referred for the interpretation of the results obtained. Each gene network clearly corresponds to a biological function in response to wounding as follows:
- Network 1 contains five genes, four of which are enzymes for biosynthesis of jasmonic acid (JA), which is the phytohormone that has a central signaling role in the wounding response. The other gene, AtMyc2, encodes a transcription factor that regulates JA-responsive genes (26). Gene expression profiles prepared in each locus page show that all of these genes are upregulated at an early stage of wounding (data not shown in Figure 1).
- Network 2 contains five genes that are enzymes for biosynthesis of flavonoid, one of the major secondary metabolites functioning as a phytoalexin or protectant against photodamage.
- Network 4 contains two JA-responsive genes. CORI3 is an enzyme, cystine CS lyase (30), which is regulated by JA signaling. This enzyme is involved in the sulfur metabolism participating in defence mechanism. Although LOX2 had been thought to act in JA biosynthesis (31), this gene is known to respond to wounding and JA stimulus at a late stage (32,33) and thus may be involved in JA synthesis at a late stage or function for other purpose in the wounding response. In this example, four groups of wounding-responsive genes are found. To make a complete list of genes related to the wounding response, next search for co-expressed genes from each group. To select genes to query, click on the checkboxes to the left of each appropriate entry—for example, the group of genes for the JA biosynthetic pathway above—and the click the button ‘search co-expressed genes’ just above the table.
The result of the co-expressed gene search is a list of the 300 most strongly co-expressed genes from the five query genes for JA biosynthesis (Figure 1D). This list contains putative functional and unknown genes in addition to many known JA biosynthesis genes. In the same way, we can obtain co-expressed genes for other gene groups. Finally, gene lists containing putative wounding-responsive genes are obtained. These lists of genes are supported by other biological information or cis-element information and can be used to design experiments to identify gene functions.
EXAMPLE 2: OBTAINING PREDICTED CIS ELEMENTS
Information of predicted cis elements can provide support for proposed co-expressed gene relationships. When co-expressed genes share particular cis elements in their promoters, we can expect that these genes are co-regulated through these cis elements and that they function together. As an example in obtaining information on predicted cis elements, consider the regulation of an abscisic acid (ABA)-responsive protein At5g62490. ABA is a phytohormone for stress response signaling. ATTED-II indicates that this gene can be up-regulated at embryogenesis through an ABA-responsive element (ABRE) (Figure 2). (Note that larger figures for this example are shown in the help page in ATTED-II.)
Search ‘At5g62490’ as an AGI code using the search form to the right of the title logo (Figure 2A).
Click the AGI code in the resulting table to go to the locus page (Figure 2B).
In the locus page for At5g62490, cis elements are predicted at 60–62 bp and 101–103 bp upstream from the TSS (Figure 2C). Although six predicted heptamers are provided, they are grouped into two cis elements for position, GACACGTGT and CCACGTGGC, sharing CACGTG core. The co-expressed gene list indicates that this gene is co-expressed with genes for late embryogenesis. Graphs of At5g62490s expression pattern are also provided, showing that this gene is upregulated by osmotic and salt stresses as well as ABA treatment (data not shown in Figure 2C). Dramatic expression during embryogenesis is also shown, suggesting that this gene may function in embryogenesis. Click ‘link to top 300 co-expressed gene list’.
In the co-expressed genes page for At5g62490, appearance of ACACGTG in the co-expressed genes is statistically abundant, suggesting that these genes, including At5g62490 itself, are commonly regulated through this cis element (Figure 2D). To obtain detailed information on this cis element, return to the locus page and click the link ‘ACACGTG’ to go to the cis element page.
In the cis element page for ACACGTG (Figure 2E), users can see that this predicted heptamer corresponds to the reported cis elements named ABRE and G-box. The ABRE is a central cis element of ABA signal transduction in response to stress treatment (34). This element prefers the position from 50 to 100 bp upstream of the TSS, indicating that the ABRE-like element found in At5g62490 appears in an appropriate position (60–62 bp upstream). The graph for cis-element specificity for various experimental conditions shows that genes containing this cis element in their promoters are usually up-regulated responding to ABA as well as osmotic or salt stress (data not shown in Figure 2). This pattern is consistent with the expression pattern of At5g62490, supporting the possibility that this gene is regulated through this cis element. A list of transcription factor candidates that bind this heptamer is also provided on this page. This list includes known transcription factors to bind ABA-responsive cis elements or G-box. In fact, ABF3, a well-known transcription factor to bind ABRE (35), is found in the list, although its correlation value is not high for this sequence itself. When we focus on the regulation during embryogenesis, RD26 (At4g27410) is a good candidate because this gene is highly expressed during embryogenesis and up-regulated by ABA as well as osmotic or salt stress, as indicated by their expression patterns on their respective locus page. Furthermore, RD26 functions in ABA-dependent stress signaling (36), and its homologous transcription factors recognize a CACG core (37). G-box binding factor 3 (GBF3, At2g46270) is another candidate, since it binds G-box sequence for ABA signaling (38). The cis element pages for the second group of heptamers found in the locus page for At5g62490 contain almost identical information except for the position preference, which is at 100 bp upstream of the TSS, indicating that the second cis element also appears at its most appropriate position (101–103 bp).
In summary, information in ATTED-II suggested the following regulatory scheme for the test case of At5g62490. Osmotic or salt stress stimulates ABA signaling. The ABA signal up-regulates At5g62490 together with co-expressed genes for late embryogenesis, potentially through two ABREs and the transcription factors RD26 and GBF3.
OTHER METHODS TO ACCESS PAGES IN ATTED-II
ATTED-II also provides lists of functional categories and cis elements to browse. Direct access by entering the URLs is also possible. For example, using http://www.atted.bio.titech.ac.jp/locus/(AGI code of the gene of interest).html will bring up the locus page of the gene of interest.
DATA SOURCES AND CALCULATION
Definition of co-expressed genes in ATTED-II
To define co-expressed genes, gene expression profiles were compared. The profiles were constructed from 1388 GeneChip data downloaded from TAIR (4), which were manually divided into 53 experimental groups and normalized by RMA method (39). To quantify the similarity of the gene expression profiles, pairwise Pearson's correlation coefficients were used (40). The 12 most strongly correlated genes for each gene (including the gene itself) were used to generate the co-expressed gene networks. The 300 most strongly correlated genes for each gene were listed in the co-expressed genes page. The lists of all co-expressed genes are downloadable from ATTED-II as tab-delimited text files. Co-expressed gene networks were pictured by simply drawing a line between co-expressed genes. Details of the method and description of the experiments used in ATTED-II are available in the help pages of ATTED-II.
Prediction of cis elements
Previous reports on comprehensive cis element prediction for Arabidopsis have used reported cis elements to search the genome (20,41) or have extracted conserved motifs from the genome (42,43). Our approach using microarray data complements their approaches. To estimate gene regulatory schemes, cis elements in the region 200 bp upstream of the TSS, which is downloaded from TAIR (4), were predicted based on the method described previously (44). Briefly, oligomers were used as cis-element candidates, and correlations between gene expression changes and presence of the cis-element candidates in the gene promoters were estimated by calculation of Pearson's correlation coefficients. This value is named CEG (correlation between gene expression and a defined gene group) in ATTED-II. We modified this method to distinguish the position of the heptamers from the TSS to reduce false positives.
The region used to extract cis elements was also examined. The peak of position specificity was concentrated within 200 bp of the TSS. Moreover, estimation of expression changes using the extracted cis elements by a linear model (44) was best for cis elements within 200 bp upstream of the TSS, indicating that most of the cis elements predicted out of this region are false positives (data not shown). These results agreed with a previous report about ABA-responsive elements (34). Additionally, the shorter region can save the calculation cost. Thus we decided to use the region 200 bp upstream from the TSS.
As a result, 304 heptamers were extracted from all 16 384 (=47) possible heptamers and registered in ATTED-II. The properties of position specificities are presented in the cis element pages. Details of our methods are described in the help pages in ATTED-II.
Acknowledgments
We thank Dr Takayuki Tohge, Mr Tetsuya Sakurai and Mr Keiji Akiyama for valuable discussions about utilization of the co-expressed gene lists. We also thank Dr Nicolas Sierro for his comments on the manuscript. This work was supported in part by the project ‘Development of Fundamental Technologies for Controlling the Production of Industrial Materials by Plants’ supported by NEDO (New Energy and Industrial Technology Development Organization, Japan) and by CREST (Core Research for Evolutional Science and Technology) of the Japan Science and Technology Agency. Computation time was provided by the Super Computer System, Human Genome Center, Institute of Medical Science, The University of Tokyo. Funding to pay the Open Access publication charges for this article was provided by Tokyo Institute of Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Parkinson H., Sarkans U., Shojatalab M., Abeygunawardena N., Contrino S., Coulson R., Farne A., Lara G.G., Holloway E., Kapushesky M., et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2005;33:D553–D555. doi: 10.1093/nar/gki056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett T., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R. NCBI GEO: mining millions of expression profiles–database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikeo K., Ishi-i J., Tamura T., Gojobori T., Tateno Y. CIBEX: center for information biology gene expression database. C. R. Biol. 2003;326:1079–1082. doi: 10.1016/j.crvi.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 4.Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craigon D.J., James N., Okyere J., Higgins J., Jotham J., May S. NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinhauser D., Usadel B., Luedemann A., Thimm O., Kopka J. CSB.DB: a comprehensive systems-biology database. Bioinformatics. 2004;20:3647–3651. doi: 10.1093/bioinformatics/bth398. [DOI] [PubMed] [Google Scholar]
- 7.Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 8.Manfield I.W., Jen C.H., Pinney J.W., Michalopoulos I., Bradford J.R., Gilmartin P.M., Westhead D.R. Arabidopsis Co-expression Tool (ACT): web server tools for microarray-based gene expression analysis. Nucleic Acids Res. 2006;34:W504–W509. doi: 10.1093/nar/gkl204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmermann P., Hennig L., Gruissem W. Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 2005;10:407–409. doi: 10.1016/j.tplants.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Lisso J., Steinhauser D., Altmann T., Kopka J., Mussig C. Identification of brassinosteroid-related genes by means of transcript co-response analyses. Nucleic Acids Res. 2005;33:2685–2696. doi: 10.1093/nar/gki566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persson S., Wei H., Milne J., Page G.P., Somerville C.R. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl Acad. Sci. USA. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rautengarten C., Steinhauser D., Bussis D., Stintzi A., Schaller A., Kopka J., Altmann T. Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput. Biol. 2005;1:e40. doi: 10.1371/journal.pcbi.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gachon C.M., Langlois-Meurinne M., Henry Y., Saindrenan P. Transcriptional co-regulation of secondary metabolism enzymes in Arabidopsis: functional and evolutionary implications. Plant Mol. Biol. 2005;58:229–245. doi: 10.1007/s11103-005-5346-5. [DOI] [PubMed] [Google Scholar]
- 14.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouze P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rougemont J., Hingamp P. DNA microarray data and contextual analysis of correlation graphs. BMC Bioinformatics. 2003;4:15. doi: 10.1186/1471-2105-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obayashi T., Okegawa T., Sasaki-Sekimoto Y., Shimada H., Masuda T., Asamizu E., Nakamura Y., Shibata D., Tabata S., Takamiya K., et al. Distinctive features of plant organs characterized by global analysis of gene expression in Arabidopsis. DNA Res. 2004;11:11–25. doi: 10.1093/dnares/11.1.11. [DOI] [PubMed] [Google Scholar]
- 18.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 19.Paul Horton K.-J.P., Obayashi T., Nakai K. Protein Subcellular Localization Prediction with WoLF PSORT. Proceedings of the 4th Annual Asia Pacific Bioinformatics Conference (APBC06); Taiwan: Taipei; 2006. pp. 39–48. [Google Scholar]
- 20.Palaniswamy S.K., James S., Sun H., Lamb R.S., Davuluri R.V., Grotewold E. AGRIS and AtRegNet. a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 2006;140:818–829. doi: 10.1104/pp.105.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berardini T.Z., Mundodi S., Reiser L., Huala E., Garcia-Hernandez M., Zhang P., Mueller L.A., Yoon J., Doyle A., Lander G., et al. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 2004;135:745–755. doi: 10.1104/pp.104.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P., Foerster H., Tissier C.P., Mueller L., Paley S., Karp P.D., Rhee S.Y. MetaCyc and AraCyc. Metabolic pathway databases for plant research. Plant Physiol. 2005;138:27–37. doi: 10.1104/pp.105.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokimatsu T., Sakurai N., Suzuki H., Ohta H., Nishitani K., Koyama T., Umezawa T., Misawa N., Saito K., Shibata D. KaPPA-view: a web-based analysis tool for integration of transcript and metabolite data on plant metabolic pathway maps. Plant Physiol. 2005;138:1289–1300. doi: 10.1104/pp.105.060525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 26.Lorenzo O., Chico J.M., Sanchez-Serrano J.J., Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F., D'Auria J.C., Tholl D., Ross J.R., Gershenzon J., Noel J.P., Pichersky E. An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 2003;36:577–588. doi: 10.1046/j.1365-313x.2003.01902.x. [DOI] [PubMed] [Google Scholar]
- 28.Seo H.S., Song J.T., Cheong J.J., Lee Y.H., Lee Y.W., Hwang I., Lee J.S., Choi Y.D. Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc. Natl Acad. Sci. USA. 2001;98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faldt J., Arimura G., Gershenzon J., Takabayashi J., Bohlmann J. Functional identification of AtTPS03 as (E)-β-ocimene synthase: a monoterpene synthase catalyzing jasmonate- and wound-induced volatile formation in Arabidopsis thaliana. Planta. 2003;216:745–751. doi: 10.1007/s00425-002-0924-0. [DOI] [PubMed] [Google Scholar]
- 30.Jones P.R., Manabe T., Awazuhara M., Saito K. A new member of plant CS-lyases. A cystine lyase from Arabidopsis thaliana. J. Biol. Chem. 2003;278:10291–10296. doi: 10.1074/jbc.M212207200. [DOI] [PubMed] [Google Scholar]
- 31.Bell E., Creelman R.A., Mullet J.E. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl Acad. Sci. USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reymond P., Weber H., Damond M., Farmer E.E. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–720. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki Y., Asamizu E., Shibata D., Nakamura Y., Kaneko T., Awai K., Amagai M., Kuwata C., Tsugane T., Masuda T., et al. Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 2001;8:153–161. doi: 10.1093/dnares/8.4.153. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W., Ruan J., Ho T.H., You Y., Yu T., Quatrano R.S. Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics. 2005;21:3074–3081. doi: 10.1093/bioinformatics/bti490. [DOI] [PubMed] [Google Scholar]
- 35.Choi H., Hong J., Ha J., Kang J., Kim S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 36.Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., Tran L.S., Yamaguchi-Shinozaki K., Shinozaki K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004;39:863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 37.Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu G., Paul A.L., McCarty D.R., Ferl R.J. Transcription factor veracity: is GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell. 1996;8:847–857. doi: 10.1105/tpc.8.5.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 40.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steffens N.O., Galuschka C., Schindler M., Bulow L., Hehl R. AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res. 2004;32:D368–D372. doi: 10.1093/nar/gkh017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janaki C., Joshi R.R. Motif detection in Arabidopsis: correlation with gene expression data. In Silico Biol. 2004;4:149–161. [PubMed] [Google Scholar]
- 43.Molina C., Grotewold E. Genome wide analysis of Arabidopsis core promoters. BMC Genomics. 2005;6:25. doi: 10.1186/1471-2164-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bussemaker H.J., Li H., Siggia E.D. Regulatory element detection using correlation with expression. Nature Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]