Abstract
ForestTreeDB is intended as a resource that centralizes large-scale expressed sequence tag (EST) sequencing results from several tree species (http://foresttree.org/ftdb). It currently encompasses 344 878 quality sequences from 68 libraries, from diverse organs of conifer and hybrid poplar trees. It utilizes the Nimbus data model to provide a hosting system for multiple projects, and uses object-relational mapping APIs in Java and Perl for data accesses within an Oracle database designed to be scalable, maintainable and extendable. Transcriptome builds or unigene sets occupy the focal point of the system. Several of the five current species-specific unigenes were used to design microarrays and SNP resources. The ForestTreeDB web application provides the means for multiple combination database queries. It presents the user with a list of discrete queries to retrieve and download large EST datasets or sequences from precompiled unigene assemblies. Functional annotation assignment is not trivial in conifers which are distantly related to angiosperm model plants. Optimal annotations are achieved through database queries that integrate results from several procedures based open-source tools. ForestTreeDB aims to facilitate sequence mining of coherent annotations in multiple species to support comparative genomic approaches. We plan to continuously enrich ForestTreeDB with other resources through collaborations with other genomic projects.
INTRODUCTION
The large sizes of conifer genomes (∼60 billion bases) make them unlikely candidates for complete genome sequencing in the immediate future. To enable the development of genomic applications in forest trees, several large-scale expressed sequence tag (EST) sequencing projects have been initiated worldwide [(1–3); http://www.treenomix.ca, http://www.pine.msstate.edu]. The major anticipated outcomes of these and other forest genomics projects involve the development of molecular applications ranging from tree breeding to eco-physiology and the design of effective conservation strategies. To enable the development of such genomic applications in forest trees, microarrays and single nucleotide polymorphism (SNP) resources have already been developed (4). The mining of expression and SNP data requires a coherent annotation of the sequence resources. Functional assignations of conifer sequences are especially challenging since these species are distantly related to angiosperm plant models for which significantly more data and tools are already available (2).
Our group at the Center for Computational Genomics and Bioinformatics (CCGB, University of Minnesota) aims to contribute to the annotation of forest tree sequences through collaborations with groups involved in forestry research. We have developed an annotation pipeline making use of several publicly available software and sequence repositories (5). We applied the annotation procedure to several EST collections obtained in conifer and poplar species. Unifying data related to several EST projects, the ForestTreeDB database is dedicated to store and handle these sequence and annotation data. The aim of this work was to produce an extensive EST database for tree species with links to other related plant resources. ForestTreeDB is a dynamic structure which will be continuously enriched with other sequence resources and new features in the future. Its purpose is to make sequence annotation available for the wide community of biologists involved in tree research, and to provide a flexible interface for developing queries. The other main publicly accessible EST databases that include forest tree species are the Gene Indices (6) and the PlantGDB database (7). Although a partial overlap exists between theses databases, each brings distinct analytical approaches. Moreover, ForestTreeDB hosts only sequences that were subjected to a stringent quality filtering. Such a procedure provides a high level of confidence in the data which is critical for analyses of gene subsequently diversity, among others.
DATA COLLECTED FROM FOREST GENOMICS PROJECTS
ForestTreeDB includes 344 878 quality sequences from loblolly pine, white spruce and poplar, derived from 243 707 cDNA clones. All in all, it currently represents EST data derived from 68 cDNA libraries produced by different projects. All of the corresponding EST sequences have been released to dbEST.
In loblolly pine (Pinus taeda L.), data collected from two projects funded through the NSF Plant Genome Research Program are represented in ForestTreeDB. The first project was developed by the group of Dr Sederoff at North Carolina State University to analyse wood development, and is very much focused on xylem cDNA sequencing (http://pine.ccgb.umn.edu/). Six libraries were prepared from differentiating xylem tissues collected from different organs (stem, root) and representing different developmental stages (juvenile wood, mature normal wood, late wood and ‘planings’ enriched for more highly differentiated xylem) or after bending of trees (compression wood, side wood). The detailed descriptions of the libraries are provided at http://pinetree.ccgb.umn.edu/documents/pine_libraries/lib_index.html. The sequences were obtained from the 5′ end of directionally cloned cDNA inserts. The other loblolly pine projects are headed by Dr Dean at the University of Georgia (UGA) (http://fungen.org/Projects/Pine/Pine.htm). This group has prepared a total of 34 cDNA libraries. Within the project entitled ‘Transcriptome responses to environmental conditions in loblolly pine roots’, libraries were prepared from roots following drought stress or various nutrient treatments (macro or micronutrients). The group also generated libraries from pine challenged with the necrotrophic fungus Fusarium circinatum, the inciting agent of pitch canker disease. Sequences were obtained from both the 3′ and 5′ ends of the inserts and 229 867 pine ESTs were generated. The Arborea project (http://www.arborea.ulaval.ca) has produced ESTs from white spruce [Picea glauca (Moench) Voss], a softwood species economically important in Canada. Gene discovery was undertaken by producing cDNA libraries from diverse spruce organs; each library representing several developmental stages, manipulative treatments and/or time points (5). In total, 17 libraries were explored from which random clones were sequenced from the 3′ end. Close to half of the clones, selected from among the best libraries, were also sequenced from the 5′ end. At present, the database contains 49 102 white spruce quality reads, and the project will sequence 150 000 additional ESTs in the coming year. Arborea also produced ESTs in poplar (Populus balsamifera subsp. trichocarpa × Populus deltoides). In this species, eight libraries of cDNAs were sequenced from the 5′ end alone, and 10 223 EST sequences have been incorporated into ForestTreeDB. Arborea sequences incorporated into the database represent recent assemblies; however, the project is continuing to add new sequences and update assemblies for future addition to the database. In addition, poplar sequences will be added from other major EST projects, including those generated for the poplar genome sequencing project.
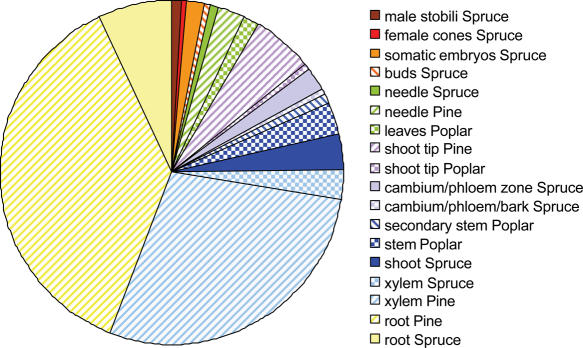
The pine and spruce sequence resource residing in ForestTreeDB represent a large proportion of the public domain data available for conifers. In contrast, the current poplar data present is a small fraction of the sequences available for this genus. The 243 707 cDNA clones represented in the database offer a broad sampling of organs encompassing numerous tissue types. Their distribution across the different organs used to prepare the 68 cDNA libraries is summarized in Figure 1. Sequences expressed in root tissues undergoing a wide diversity of chemical or environmental treatments sampled in pine and spruce represent 44.2% of the sequence database. The second largest sample include 41% of the sequences found in libraries from stems and vascular tissues in pine, spruce or poplar. Sequences from poplar leaves and conifer needles account for 4.7% of the overall sequence data. The remaining sequences were derived from cDNA libraries prepared from various organs including cones, strobili, buds, embryos and shoot tips.
Figure 1.
Tissue sampling. Number of clones representing the different organs from pine, spruce or poplar used to prepare the cDNA libraries. The full description of the 68 cDNA libraries is available in the database in the Summaries section, Library Descriptions (http://foresttree.org:8680/DB/nimbus/query.do?action=query&query=All+Library+Names).
SEQUENCE PROCESSING AND ANALYSIS
Before entering the sequences into the database, they are processed starting from the trace files following a procedure developed at CCGB for base-calling, vector-trimming and removal of chimeric sequences. This procedure assures that only quality reads are incorporated in the database. In total, 344 878 quality sequences were derived from these clones. Processed ESTs are then assembled using Phrap [http://www.phrap.org, (8,9)]. The quality control procedure is detailed in Pavy et al. (5).
The database currently hosts five unigene sets that represent a total of 179 300 unigene sequences. The CCGB produced four of the unigene assemblies (two from white spruce and one each from loblolly pine and hybrid poplar); whereas, the fifth assembly was generated at UGA (loblolly pine). Thus, for loblolly pine, two separate unigene sets were prepared from different sequencing projects and following somewhat different procedures. A first set was derived from all the pine ESTs generated by the NSF genomics project studying wood formation in Loblolly pine (2), and was prepared at the CCGB. The other set of 122 079 loblolly pine contigs was assembled from 173 070 ESTs generated by Dean's laboratory at the University of Georgia. As can be seen from the comparison of the number of ESTs with the number of unigenes, these data were assembled with a different stringency, based upon the needs of the Georgia sequencing effort, and was incorporated as their assembly at the request of our Georgia collaborators. With white spruce, two assemblies were chosen from those produced in the Arborea sequencing project (release 7 and 8), as they serve as references for key resources developed during the project. One spruce unigene (release 7) was used to design a first generation Arborea spruce cDNA microarray (http://www.arborea.ulaval.ca). The other spruce unigene was analysed to describe the functional annotations of the targeted genes (5), to generate a candidate SNP resource (10) and to generate a second generation spruce microarray. The Arborea poplar ESTs assembly was also used to design the Arborea poplar microarrays.
We compared the conifer unigene sets against each other by using the blastn program to determine the overlap between these sequence datasets (Table 1). These comparisons have shown the complementarity of the two pine unigene sets. The two pine assemblies share 53.45% of sequences with an overlap of >80% of identity over >100 nt. Using the same similarity parameters, the pine_NSF and the pine_UGA overlap 53.45 and 66.25% of the spruce unigenes, respectively. Sequences belonging to several unigene sets can be mined simultaneously based on GO terms describing them. Sequence analyses conducted by using several approaches have been uploaded in the database.
Table 1.
Number of unigenes found in the three major conifer UnigeneSets and number of unigenes sharing 80% of identity over at least 100 nt, in pairwise comparisons
| UnigeneSet | Spruce_Arborea_Release8 | Pine_NSF | Pine_UGA |
|---|---|---|---|
| Total number of unigenes | 16 602 | 20 483 | 122 079 |
| Pine_NSF | 8874 (53.45%) | ||
| Pine_UGA | 12 470 (75.1%) | 13 571 (66.25%) |
Only one of the two spruce unigene is presented here because they are largely overlapping.
Sequence similarities were detected using the blast program (11) against sequences from UniProt/UniRef100 and the Arabidopsis TAIR resource (12), hidden Markov model (HMM) searches (13) are performed against Pfam (14), TIGRFAMS (15), SUPERFAMILY (16) and SMART motifs (17). The resulting sequence similarity results were used to correlate the contigs to terms from the Gene Ontology (GO) (5). We estimate that the process of correlating GO terms with unigenes enabled the assignment of tentative annotations to 3.4–10% of the unigenes depending on the UnigeneSet and the GO category (Table 2). A GO summary is precompiled and directly accessible through ForestTreeDB. The dedicated link displays a table providing for each GO term, the GO accession, the GO definition, the number of unigeneIDs related to this GO class, whatever the E-value associated with this functional assignation, and a link to a page that will further display the composition of these related unigenes.
Table 2.
Compilation of some statistics about sequence annotation extracted from ForestTreeDB with the query ‘GO Accn-BLAST’ in combination with a condition on the UnigeneSetID
| UnigeneSet | Species | Number of unigenes | Unigenes associated with the GO with a P-value < 1E−10 | Unigenes with a match found with P-value < 1E−10 in | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Category molecular function | Category biological process | Category cellular component | Uniref100 (blast× hit) | PFAM (HMM hit) | SUPERFAMILY (HMM hit) | TIGRFAM (HMM hit) | SMART motifs (HMM hit) | |||
| Spruce | Spruce | 16 602 | 1429 (8.6%) | 1550 (9.3%) | 771 (4.6%) | 10 205 (61.5%) | 7230 (43.5%) | 5783 (34.8%) | 12 016 (72.4%) | 10 730 (64.6%) |
| NSF_pine | Pine | 20 483 | 1880 (9.2%) | 2055 (10%) | 1022 (5%) | 6896 (33.7%) | 8687 (42.4%) | 4397 (21.5%) | 15 547 (75.9%) | 4924 (24%) |
| UGA_pine | Pine | 122 079 | 7548 (6.2%) | 8291 (6.8%) | 4203 (3.4%) | 82 261 (67.4%) | 108 825 (89.1%) | 63 963 (52.4%) | 106 548 (87.3%) | 87 173 (71.4%) |
| Arborea poplar | Poplar | 5911 | 536 (9.1%) | 585 (9.9%) | 317 (5.4%) | 4243 (71.8%) | 4393 (74.3%) | 2637 (44.6%) | 5025 (85%) | 3165 (53.5%) |
Matches were filtered out based on the E-value threshold of 1E−10. Queries were completed with the following GO accessions: GO:0005575 (cellular component), GO:0003674 (molecular function) and GO:0008150 (biological process). The number of Unigenes annotated following a blast search or HMM search against several protein databases are indicated.
The GO annotations were complemented with blast or HMM searches against several protein or motif databases (see above). This significantly augmented the number of annotated unigenes compared with the GO assignation alone (Table 2). The number of annotated unigenes was the highest for the poplar UnigeneSet; this result was expected since poplar is more closely related to Angiosperm model species, which are more prevalent in the core sequence databases. Indeed, in this unigene set >71% of the sequences were found with a match in Uniref100 (P-value < 1E−10). With the conifer unigene sets, our ability to assign sequence annotation was lower and varied significantly between the datasets. For example, 61.5% of the spruce unigenes (assembled with the same procedures as the poplar sequences) gave significant hits with blastx, whereas HMM searches produced 64.6% hits against SMART motifs and 72.4% hits against TIGRFAM (P-value < 1E−10) (Table 2). For each Unigene, a specific page displays annotation results obtained from several analyses and extensively linked to external resources. First, similarity results detected using blastx and based on HMM searches are parsed to display possible similar sequences found in other databases, and are displayed in separate tables. For each blast match, the output includes the location of the best high scoring pair, accession and description of the match, E-value, length and bit score of the alignment. In contrast to other transcriptome databases (e.g. the Gene Indices), we have not only loaded the best matches, but also all of them. We have incorporated the sequence similarity level as a parameter that the user can use to personalize the query. Second, this page lists all the GO terms inferred based on these similarity results. For each GO term, a short description is provided as well as a web link to the QuickGO page at EMBL-EBI which gives access to the complete description of the GO category.
The annotation procedures combined with the queries available through the interface is well suited to search for candidate genes, a preliminary step which is crucial before performing genetic analyses. ForestTreeDB's strength is to facilitate this procedure in a user-friendly manner. Also, as the user simultaneously combines several queries, the database interface enables flexibility for the mining of candidate genes. Finally, ESTs composing the Unigene are listed as well as the clone names they are derived from EST sequences and their alignment in an MSF format can be downloaded. To graphically assess the quality of the EST assembly, an alignment image is provided that displays the sequences in a colour-coded manner.
DATABASE
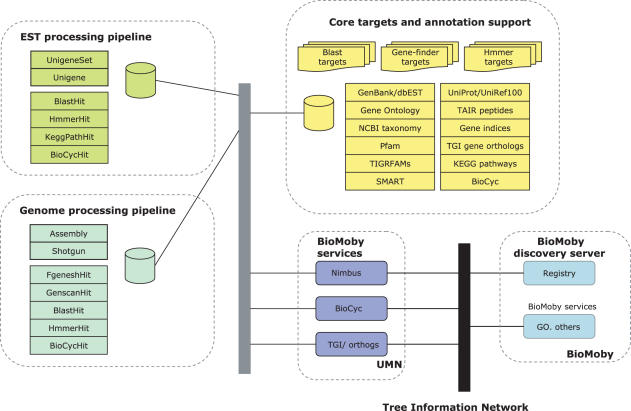
The Nimbus data model provides a hosting system for multiple EST projects, similar in Intent to the Gene Indices. From the data standpoint, each project retains Information on experimental data (libraries, clones and reads), processed results (finished sequences, assemblies) and annotations of the unigenes (e.g. blast and HMMER reports) obtained by assembly (Figure 2). Transcriptome builds occupy the focal point of the system, and information linkages between the unigenes of a build, the original experimental data, and the processed results provide a very rich representation of the transcriptome.
Figure 2.
Database environment. EST and genomic resources access the same core targets and annotation support and semantic web services [semantic BioMoby] provide the access mechanism.
Experimental information includes library name, taxon and sequenced end. A build of the transcriptome produces a set of contigs from which are selected the unigene sets. BLAST and HMMER generated annotations of each unigene may be retained; likewise, available SNP predictions may be retained. An important feature of the BLAST and HMMER hits is that the project retains only confidence levels, scores, etc.; actual reference information about the hit sequence is retained in a separate database shared by all projects. Thus instead of storing a ‘defline’ for each sequence hit by a unigene as is common, a pointer to a shared reference database is stored. The shared database provides functional and structural information on each hit in terms of its definition and known aspects using controlled vocabularies such as the Gene Ontology Consortium's. The net result is a powerful system which can be used to identify, for example, all unigenes involved in DNA binding that are unique to a particular library.
Information is stored in Oracle databases, and the Nimbus system provides object-relational mapping APIs in Java and Perl for data accesses. The Perl API is used primarily for performing bulk imports and database management. The Java API supports tiered-architecture access; it is used by the online web tools, and can be leveraged by other webapps. For example, it has been used to support semantic BioMoby services created for the Tree Information Network program (Figure 2). The web tools are developed using standard Java technologies, and can be deployed under a Tomcat server.
QUERIES AND INTERFACE
The ForestTreeDB portal, http://foresttree.org/ftdb, provides access to all the data. Two menus enable the retrieval of project summaries or allow the performance of queries and retrieval of the resulting sequences. The web interface allows multiple queries to be chained together, since the results are combined using Boolean operations. This provides the database user with the ability to create project summaries, and to identify transcripts by putative function. An example is illustrated on Figures 3 and 4. To assist the user in mining the annotation data, we provide a page describing the protocols, parameters used to run the software as well as guidelines to perform the queries.
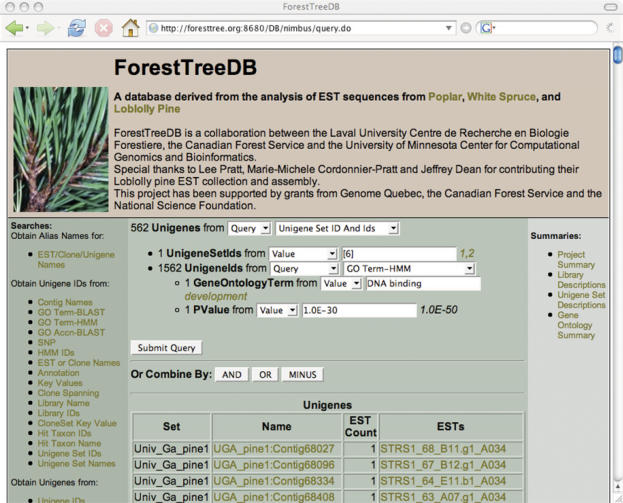
Figure 3.
ForestTreeDB screenshot showing a query combining a GO search and a specific UnigeneSet search. Unigenes were searched belonging to the UnigeneSetID 6 (pine unigenes derived from the UGA assembly) and correlated to the GO term ‘DNA binding’ with P-value < 1E−10. The query resulted in 562 Unigenes.
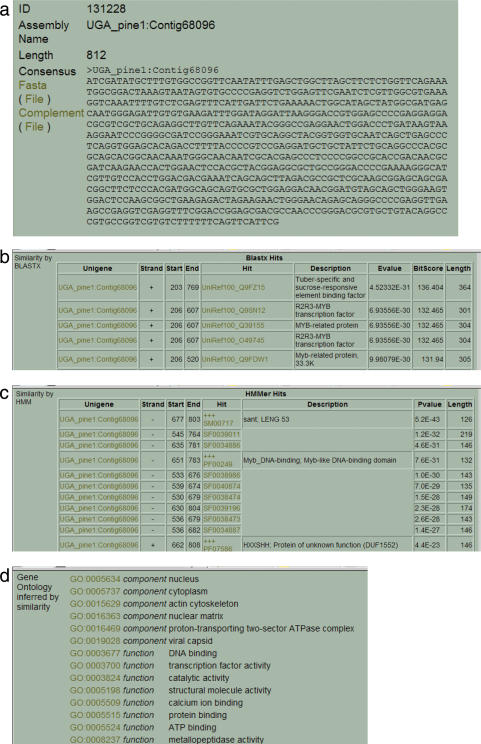
Figure 4.
ForestTreeDB screenshot displaying the annotation assigned to one of the unigene retrieved with the query from Figure 3. For each annotation method, all the matches are displayed. Here, only the top of the screenshot is shown for each query (to limit the size of the figure). (a) Contig information including identifier, sequence, links to retrieve the sequence. (b) Blastx matches including hit's accession, hit location, similarity parameters. (c) Hits found by HMMER, including accession and description of the hit, location match and similarity parameter. (d) List of terms from Gene Ontology inferred to this contig.
The ForestTreeDB web application provides the means to query the database without requiring the user to be knowledgeable in database query languages (e.g. SQL). It presents the user with a list of discrete queries. The user can select one of these queries, and the web application will build an input form to allow the user to enter input parameters for the query. The form also allows the user to combine the selected query with other queries. These queries are not hard-coded into the web application, so new queries may be easily added as the database changes and additional information is added.
These queries are entered by an administrator, knowledgeable in the ForestTreeDB schema and in database query languages. The queries are categorized by the type of data they return. For example, a number of queries may return the IDs of unigenes. The results of these queries can be combined using set operations, specifically ‘and’, ‘or’ and ‘difference’. Furthermore, the input parameters to the queries are similarly categorized so that the output of a query can be used as input to the parameter of another query; thus, complex and varied queries can be built by the user (Figure 3). These queries are more complex than those that can be provided by pre-determining and hard-coding the information that can be queried by the user.
There are several entry points to mine the database, including sequence identifiers, homolog's accession or definition, or SNPs. Sequences are referenced by the UnigeneID they belong to, the corresponding contig name in the bioDATA repository (http://biodata.ccgb.umn.edu/), names of the clone they are derived from and the accession of the ESTs deposited in dbEST. All these aliases can be obtained either from the EST or clone name. A user can start a search based on EST accession number derived from a blast query made elsewhere, look for the UnigeneIDs including this EST sequence and then mine the assigned functional annotations (Figure 4b–d). Another database entry point is a function-centred query, which can be performed in various fashions. For example, queries can be based in GO accession numbers or GO terms. The annotations may be mined based on a keyword search among the GO terms correlated with the ForestTreeDB unigenes and derived either by blast or HMM search. Furthermore, the query can be performed after selecting UnigeneSets or libraries to be included in the output.
The interface enables the user to download large datasets such as all the ESTs derived from one or several libraries, or the contigs and singletons resulting from one of the precompiled assemblies. Each query can target the libraries individually, a combination of libraries, a specified UnigeneSet or a series of UnigeneSets. Thus, it is possible to mine sequences derived from a single species or from several species in the same query. Once a query has been sent to ForestTreeDB, the number and a complete list of unigenes complying with the input parameters are returned. There is the opportunity to download files containing the sequence (Fasta format) (Figure 4a) for a single unigene or for all of the unigenes identified by the query, which facilitates other analyses and the design of target laboratory experiments. A summary table also provides links to related information residing either in ForestTreeDB or in other databases used in our annotation process.
FUTURE DEVELOPMENTS
In addition to adding new resources and analyses, a current area of development includes the development of the Tree Information Network. This work will involve the development of semantic BioMoby web services as part of an effort to make the data from this project available in a larger inter-operable framework. Services to be developed under this project include both query services, as well as some application services.
Acknowledgments
This work was supported by Genome Québec, Genome Canada for the Arborea project to J.M., and the National Science Foundation Plant Genome Research Program, and the USDA Cooperative State Research, Education and Extension Service Plant Genome Program to E.R. Funding to pay the Open Access publication charges for this article was provided by Genome Québec and Genome Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.Allona I., Quinn M., Shoop E., Swope K., St Cyr S., Carlis J., Riedl J., Retzel E., Campbell M., Sederoff R., et al. Analysis of xylem formation in pine by cDNA sequencing. Proc. Natl Acad. Sci. USA. 1998;95:9693–9698. doi: 10.1073/pnas.95.16.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirst M., Johnson A.F., Baucom C., Ulrich E., Hubbard K., Staggs R., Paule C., Retzel E., Whetten R., Sederoff R. Apparent homology of expressed genes from wood-forming tissues of loblolly pine (Pinus taeda L.) with Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2003;100:7383–7388. doi: 10.1073/pnas.1132171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorenz W.W., Sun F., Liang C., Zhao X., Kolychev D., Wang H., Cordonnier-Pratt M.-M., Pratt L.H., Dean J.F.D. Water stress-responsive genes in loblolly pine (Pinus taeda L.) roots identified by analyses of expressed sequence tag libraries. Tree Physiol. 2006;26:1–16. doi: 10.1093/treephys/26.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Ralph S., Park J.-Y., Bohlmann J., Mansfield S.D. Dirigent Proteins in Conifer Defense: gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.) Plant Mol. Biol. 2006;60:21–40. doi: 10.1007/s11103-005-2226-y. [DOI] [PubMed] [Google Scholar]
- 5.Pavy N., Paule C., Parsons L., Crow J.A., Morency M.J., Cooke J., Johnson J., Noumen E., Guillet-Claude C., Butterfield Y., et al. Generation, annotation, analysis and database integration of 16,500 white spruce EST clusters. BMC Genomics. 2005;6:144. doi: 10.1186/1471-2164-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y., Tsai J., Karamycheva S., Pertea G., Sultana R., Antonescu V., Chan A., Cheung F., Quackenbush J. The TIGR Gene Indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res. 2005;33:D71–D74. doi: 10.1093/nar/gki064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong Q., Lawrence C.J., Schlueter S.D., Wilkerson M.D., Kurtz S., Lushbough C., Brendel V. Comparative plant genomics resources at PlantGDB. Plant Physiol. 2005;139:610–618. doi: 10.1104/pp.104.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 9.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred.I Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 10.Pavy N., Parsons L., Paule C., MacKay J., Bousquet J. Automated SNP detection from a large collection of white spruce expressed sequences: contributing factors and approaches for the categorization of SNPs. BMC Genomics. 2006;7:174. doi: 10.1186/1471-2164-7-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 12.Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 14.Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haft D.H., Selengut J.D., White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gough J., Karplus K., Hughey R., Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 17.Letunic I., Copley R.R., Pils B., Pinkert S., Schultz J., Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]